Glenn Lockwood writes that the world of high-throughput sequencing is becoming increasingly dependent on HPC, and many of the problems being solved in genomics and bioinformatics are stressing aspects of system architecture and cyberinfrastructure that haven’t gotten a tremendous amount of exercise from the more traditional scientific domains in computational research.
Take, for example, the biggest and baddest DNA sequencer on the market: over the course of a three-day run, it outputs around 670 GB of raw (but compressed) sequence data, and this data is spread out over 1,400,000 files. This would translate to an average file size of around 500 KB, but the reality is that the file sizes are a lot less uniform:
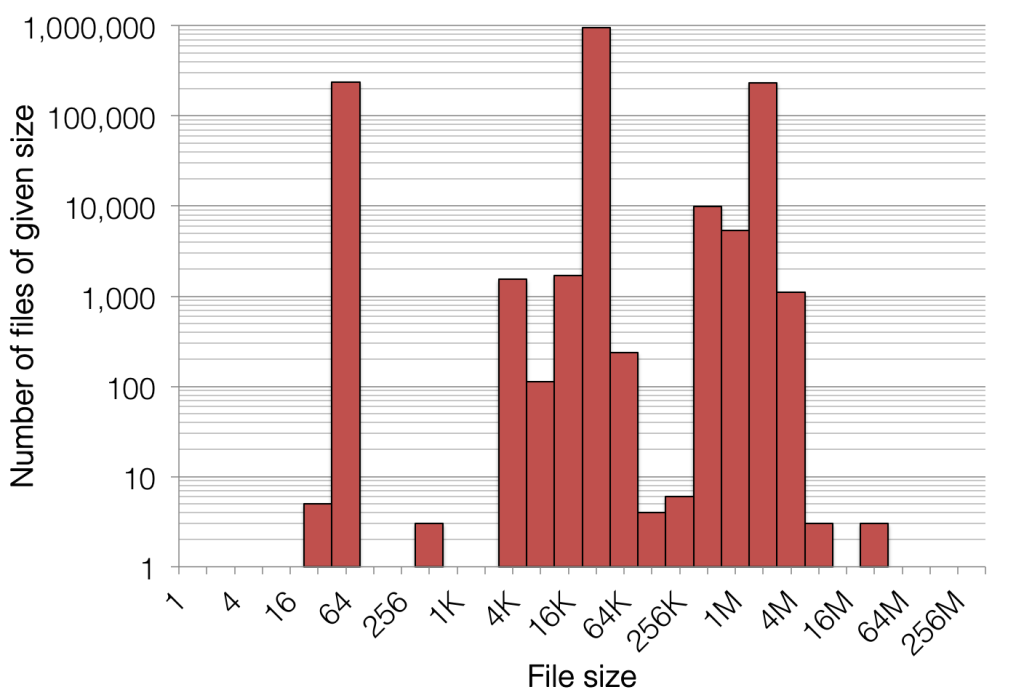
Figure 1. File size distribution of a single flow cell output (~770 gigabases) on Illumina’s highest-end sequencing platform
After some basic processing (which involves opening and closing hundreds of these files repeatedly and concurrently), these data files are converted into very large files (tens or hundreds of gigabytes each) which then get reduced down to data that is more digestible over the course of hundreds of CPU hours. As one might imagine, this entire process is very good at taxing many aspects of file systems, and on the computational side, most of this IO-intensive processing is not distributed and performance benefits most from single-stream, single-client throughput.
Read the Full Story.
Download the insideHPC Guide to HPC Storage





