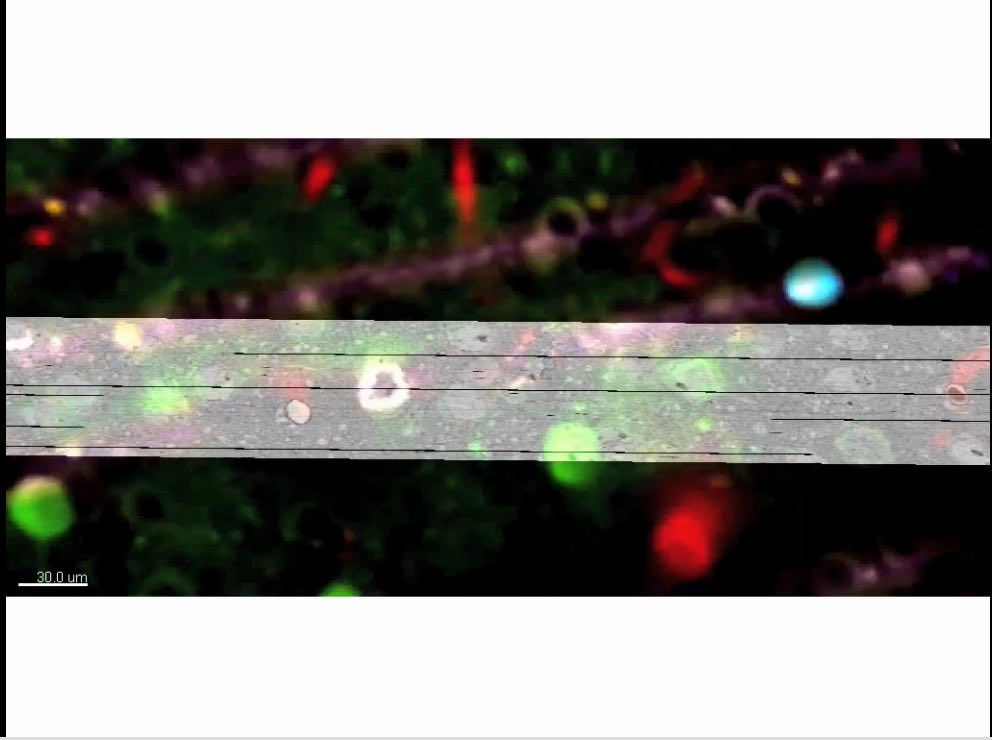
In vivo fluorescence to EM correspondence
With help from the Pittsburgh Supercomputer Center, an international team of researchers has published the largest network to date of connections between neurons in the cortex, where high-level processing occurs, and have revealed several crucial elements of how networks in the brain are organized. The results are published this week in the journal Nature.
A team led by Wei-Chung Allen Lee of Harvard University and R. Clay Reid of the Allen Institute for Brain Science in Seattle identified brain nerve cells that respond to visual elements in living mice. Then they took a series of millions of microscope images of ultra-thin (about 40-nanometer-thick) tissue slices around these nerve cells. In a collaboration facilitated by PSC’s Art Wetzel, PSC computational scientist Greg Hood helped them to reconnect these images into a three-dimensional volume using PSC’s AlignTK software. But because these slices are fragile, microscopic tears and other artifacts happened, requiring manual intervention to correct. So researchers had to move back and forth between computation and manual “repair” of the images until the quality of the aligned volume was good enough to trace the connections between the nerve cells. The team reported in the journal Nature on March 28, 2016 that mammalian excitatory nerve cells that respond to a given visual feature do indeed make more and larger connections to other excitatory nerve cells that are tuned to respond to similar visual elements.”
A network of cortical neurons whose connections were traced from a multi-terabyte 3D data set. The data were created by an electron microscope designed and built at Harvard Medical School to collect millions of images in nanoscopic detail, so that every one of the “wires” could be seen, along with the connections between them. Some of the neurons are color-coded according to their activity patterns in the living brain. This is the newest example of functional connectomics, which combines high-throughput functional imaging, at single-cell resolution, with terascale anatomy of the very same neurons.
A network of cortical neurons whose connections were traced from a multi-terabyte 3D data set. The data were created by an electron microscope designed and built at Harvard Medical School to collect millions of images in nanoscopic detail, so that every one of the “wires” could be seen, along with the connections between them. Some of the neurons are color-coded according to their activity patterns in the living brain. This is the newest example of functional connectomics, which combines high-throughput functional imaging, at single-cell resolution, with terascale anatomy of the very same neurons. Image credit: Clay Reid, Allen Institute; Wei-Chung Lee, Harvard Medical School; Sam Ingersoll, graphic artist
This is a culmination of a research program that began almost ten years ago. Brain networks are too large and complex to understand piecemeal, so we used high-throughput techniques to collect huge data sets of brain activity and brain wiring,” says R. Clay Reid, M.D., Ph.D., Senior Investigator at the Allen Institute for Brain Science. “But we are finding that the effort is absolutely worthwhile and that we are learning a tremendous amount about the structure of networks in the brain, and ultimately how the brain’s structure is linked to its function.”
Lee and Bonin began by identifying neurons in the mouse visual cortex that responded to particular visual stimuli, such as vertical or horizontal bars on a screen. Lee then made ultra-thin slices of brain and captured millions of detailed images of those targeted cells and synapses, which were then reconstructed in three dimensions. Teams of annotators on both coasts of the United States simultaneously traced individual neurons through the 3D stacks of images and located connections between individual neurons.
Analyzing this wealth of data yielded several results, including the first direct structural evidence to support the idea that neurons that do similar tasks are more likely to be connected to each other than neurons that carry out different tasks. Furthermore, those connections are larger, despite the fact that they are tangled with many other neurons that perform entirely different functions.
“Part of what makes this study unique is the combination of functional imaging and detailed microscopy,” says Reid. “The microscopic data is of unprecedented scale and detail. We gain some very powerful knowledge by first learning what function a particular neuron performs, and then seeing how it connects with neurons that do similar or dissimilar things.
“It’s like a symphony orchestra with players sitting in random seats,” Reid adds. “If you listen to only a few nearby musicians, it won’t make sense. By listening to everyone, you will understand the music; it actually becomes simpler. If you then ask who each musician is listening to, you might even figure out how they make the music. There’s no conductor, so the orchestra needs to communicate.”
This combination of methods will also be employed in an IARPA contracted project with the Allen Institute for Brain Science, Baylor College of Medicine, and Princeton University, which seeks to scale these methods to a larger segment of brain tissue. The data of the present study is being made available online for other researchers to investigate.