 Researchers at Purdue University are using HPC to help fight the Zika virus.
Researchers at Purdue University are using HPC to help fight the Zika virus.
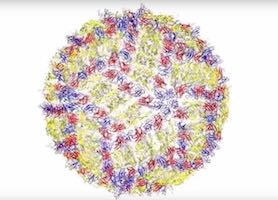
Purdue’s award-winning Community Cluster Program played a significant role in enabling the research team to create the first detailed, 3-D structural map of the Zika virus, a key step toward developing treatments. The technique the team used to map the virus structure combined cryo-electron microscopy and high-performance computing. With the microscope, the Purdue researchers capture images of many Zika virusDetailed image of the Zika virus structure. particles, or individual instances of the virus. They then use Purdue’s community cluster supercomputers to turn those images into a unified, finely detailed picture of the overall virus structure, opening a window on how it works.
“When we built the first community cluster in 2008, we knew the demand was there among our faculty researchers in a number of fields, for example in engineering and physical sciences,” says Donna Cumberland, executive director for research computing. “While Zika virus wasn’t on the radar then, we also could see the breadth and variety of research being done on the clusters today coming in the future.”
The Zika structure is the latest in a continuing series of notable research enabled by the Community Cluster Program. Each year since 2008 Purdue has built a research cluster, with seven of them ranked in the top 500 in the world, and another planned this fall. The program gives Purdue’s faculty the best collection of high-performance computing resources for use by researchers on a single campus in the country.
There are now more than 180 faculty partners and hundreds of researchers from their labs in all of Purdue’s primary colleges and schools using the community clusters for research spanning more than 30 science, engineering and social science disciplines.
Little is known about Zika virus — yet. With its structure mapped, researchers can begin to identify potential targets for treating the virus or vaccines against it. The virus structure could not have been mapped, not in timely fashion anyway, without a tool like the community clusters.
We have tens of thousands of pictures of the Zika virus and each one is in a different, random orientation,” says Michael Rossmann, Purdue’s Hanley Distinguished Professor of Biological Sciences. “If we can find out the relative orientations of all those thousands of particles, then we can put those together to make a three-dimensional image.”
High-performance computing — in this instance Purdue’s Snyder community cluster, designed for memory-intensive life science research — is employed first to whittle down the collection to the best images, from 60,000 to 10,000 in the case of Zika. Next, the computer links similarly oriented views of the virus to each other, kind of like organizing pieces of a very large puzzle. The cluster then assembles those puzzle pieces into a 3-D image of the virus structure at near-atomic resolution.
“Then, if we’re interested in how the virus can be inhibited, neutralized, we have to do many structures, each one of which is like that, of the virus complex interacting with antibodies,” says Rossmann, who led the research team with Richard Kuhn, director of the Purdue’s Institute for Inflammation, Immunology and Infectious Diseases.
Purdue’s goal is to have global impact and the Zika research wasn’t the first time our high-performance computing capability has been an integral part of doing that, nor will it be the last,” says Gerry McCartney, Purdue’s CIO and vice president for information technology. “We planned to be a leader in enabling research driven by HPC when we started the Community Cluster Program, and we plan to continue to be.”
Source: Purdue



