A workflow to support genomic sequencing requires a collaborative effort between many research groups and a process from initial sampling to final analysis. Pre-processing steps involve:
- Sampling and isolating the extracted DNA/RNA of an organism (e.g. a mouse or wheat) in an intensive and specialized library preparation process
- Loading the DNA/RNA onto the sequencing instrument
- Executing a sequencing process that, depending on the platform and preparation, may be run for days generating hundreds of GBs (or more) of data per day for a single human genome
- Performing data clean-up operations that will reduce the data size in the range from hundreds of GBs to tens of GBs per genome.
While these data pre-processing operations are effectively simple string analysis problems that can typically be run on standard Linux nodes, the sequence assembly that follows is significantly more compute and memory intensive. The assembly process may take days to weeks on low-memory cluster nodes as data make their way between various processing nodes and the centralized storage that supports the overall environment.
As can be seen in Figure 2, a typical genomics workflow diagram, the traditional cluster-based processing model involves repeatedly moving large data sets to and from a centralized storage platform; this can lead to significant delays and process inefficiencies.
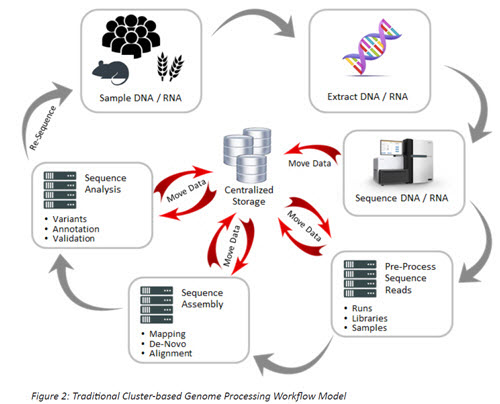
While the traditional approach to processing will certainly deliver results, the emergence of new compute and storage technologies provides opportunities for significant workflow acceleration through in-memory processing and through the use of a storage model that eliminates most of the data movement inherent in the traditional workflow. This converged model for compute and storage is based on a single image system (or scale-up system) that combines a large number of compute cores closely coupled to a high-performance flash-based storage system – sometimes called in-situ processing – that is backed by a policy-based high-capacity data management system for long term storage.
To learn how genomic sequencing is enabling personalized medicine download the insideHPC Guide to Personalized Medicine and Genomics, courtesy of SGI and Intel.




