Linding Lab at the University of Copenhagen used an SGI UV server system to discover how genetic diseases such as cancer systematically attack the networks controlling human cells. By developing advanced algorithms to integrate data from quantitative mass-spectrometry and next generation sequencing of tumor samples, the UCPH researchers have been able to uncover cancer related changes to phospho-signaling networks at a global scale. The studies are some of the early results of the strategic collaboration between SGI and the Linding Lab at UCPH. The landmark findings have been published in two back-to-back papers in today’s Cell journal.
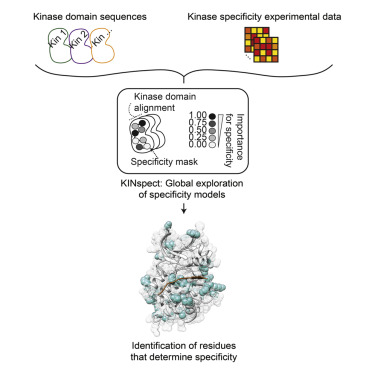
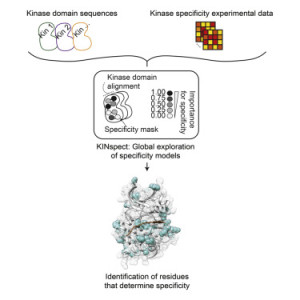
This new breakthrough allows researchers to identify the effects of mutations on the function of proteins in cancer for individual patients, even if those mutations are very rare,” said Professor Dr. Rune Linding, lead researcher on the projects from the Biotech Research & Innovation Centre, UCPH. “The identification of distinct changes within our tissues that help predict and treat cancer is a major step forward and we are confident it can aid in the development of novel therapies and screening techniques. In these studies we simulated more than 2.5 million different computer models to find the optimal parameters to interpret cancer genomes. This is a vast computational and big data challenge that requires an extreme degree of computational flexibility.”
Since the human genome was sequenced more than a decade ago, cancer genomics studies have dominated the life sciences worldwide and have been extremely successful at identifying mutations in individual patients and tumors. However, using this knowledge to develop improved cancer therapies has been severely hampered by the inability of researchers to explain and relate this data to proteins: the targets of most pharmaceutical drugs.
Using the SGI UV server platform and Intel Xeon processors, researchers from the Universities of Copenhagen, Yale, Zurich, Rome, and Tottori (Japan) have unraveled how mutations such as those acquired in cancer, target and damage the protein signaling networks within human cells on an unprecedented scale.
 The studies highlight the importance of big data in cancer biology and underpin the essentiality of large dynamic-range computing platforms such as the SGI UV. SGI’s UV server platform offers unique capabilities for research computing, well beyond what is commonly possible with commodity computing hardware. The SGI UV line combines industry-leading shared-memory designs with unmatched data performance capabilities, making it the ideal choice for big data research workflows.
The studies highlight the importance of big data in cancer biology and underpin the essentiality of large dynamic-range computing platforms such as the SGI UV. SGI’s UV server platform offers unique capabilities for research computing, well beyond what is commonly possible with commodity computing hardware. The SGI UV line combines industry-leading shared-memory designs with unmatched data performance capabilities, making it the ideal choice for big data research workflows.
There is going to be more and more data available to us, and as scientists trying to lower the cancer burden, technology like SGI’s UV system can make sense of all this data. This technology is a real game changer and these findings are a significant discovery from life sciences using a supercomputer, which we hope will make a difference for cancer patients world-wide,” continued Linding.
The interpretation of these big datasets requires more advanced modeling frameworks than traditional bioinformatics approaches. In particular, models need to account for the inherent variability and heterogeneity of biological data, which can only be achieved in a rigorous manner by probabilistic Bayesian methodologies. As these methods in turn are much more computationally demanding, technologies like the SGI UV system are becoming mandatory to support scientific analysis and creativity, and to advance our understanding of, and ability to treat, complex diseases such as cancer.
Thanks to the power of the technology in our supercomputers, SGI supports a broad range of fascinating and history-making research projects that will leave a strong mark in the life sciences and on the medical science community,” said Jorge Titinger, president and CEO, SGI. “We are honored to be a part of such a monumental research program and are looking forward to continuing to provide the computing power the Linding Lab requires to dive deeper into understanding cancer through genomic research.”
The two studies are available today in an advanced online publication and will be printed in the September 24th issue of Cell. More information about the studies and links to media content can be found on http://www.lindinglab.science and http://www.bric.ku.dk. The work was supported by the European Research Council (ERC), the Lundbeck Foundation and Human Frontier Science Program.