
insideHPC Guide to Personalized Medicine and Genomics – Download
The insideHPC Guide to Personalized Medicine and Genomics explains how genomics will accelerate personalized medicine including several case studies. Here is the introduction to this white paper.
If the keys to health, longevity, and a better overall quality of life are encoded in our individual genetic make-up then few advances in the history of medicine can match the significance and potential impact of the Human Genome Project. Instigated in 1985 and launched in 1990, it was announced as completed two years ahead of schedule in 2003 – and at a cost of over $3 billion. It delivered a 99% complete reference human genome sequence at high accuracy, but this was based on a small number of anonymous European donors. Since that time, the race has been centered on dramatically improving the breadth and depth of genomic understanding – each region wants its own reference genome sets – as well as reducing the costs involved in sequencing, storing, and processing an individual’s genomic information.
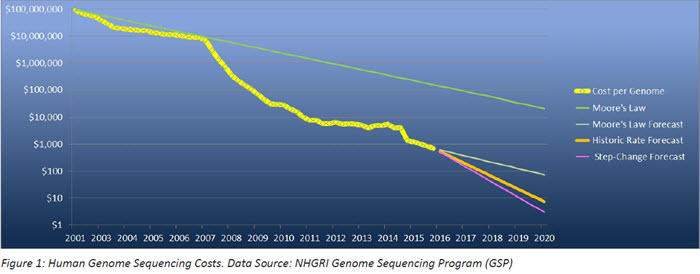
That race has been paying off. Just a little more than twenty years later, advances in computer technology have reduced the cost of sequencing an individual’s genome from around $100M to around $1,000 currently – five orders of magnitude. This is projected to decrease by at least another order of magnitude to $100 or possibly even lower than $10 per genome by the end of the decade.
This drastic reduction in sequencing costs does, however, raise many more significant problems as the overwhelming volume of data being generated accelerates. The requirement to store and process all of that information only increases as the costs of sequencing fall still further. Starting from the original European-based reference genome, many more regionally diverse reference and individual genomes are being sequenced to support populations around the world including countries in Asia, Africa, the Americas, Europe, Russia, and more. Initially, these projects are generating hundreds of thousands of genomes per year and rising to millions of genomes per year by the end of the decade. By 2030, this will amount to hundreds of millions of genomes, each at approximately half a terabyte of raw image data per human genome. These files are complex as they encompass highly granular, unstructured scientific data which is difficult to manage and analyze. Collectively, they will add up to tens or hundreds of exabytes of data. By comparison, the typical modern PC or laptop could only hold the data for a single genome at best, let alone do anything useful with it.
The benefits of personalized medicine – more precise, predictable, and powerful health care that is customized for each individual patient – are huge, but the data management challenges are extremely daunting. Genomics data management and processing brings together big data and analytics which for a long time has been the domain of High Performance Computing (HPC) disciplines. Scientists engaged in academic, clinical, and pharmaceutical research are being deluged by an expanding ‘ocean of data’ generated by sequencing. Storing, managing, accessing, analyzing, and interpreting that huge volume of data requires a new workflow model that brings storage and computing power much closer together in order to minimize the lost productivity that occurs when large amounts of data need to be repeatedly moved around to support analysis operations.
To learn how genomics is enabling personalized medicine download the insideHPC Guide to Personalized Medicine and Genomics, courtesy of SGI and Intel.




