 Using a unique computational approach to rapidly sample proteins in their natural state of gyrating, bobbing, and weaving, a research team from UC San Diego and Monash University in Australia has identified promising drug leads that may selectively combat heart disease, from arrhythmias to cardiac failure.
Using a unique computational approach to rapidly sample proteins in their natural state of gyrating, bobbing, and weaving, a research team from UC San Diego and Monash University in Australia has identified promising drug leads that may selectively combat heart disease, from arrhythmias to cardiac failure.
Reported in the September 5, 2016 Proceedings of the National Academy of Sciences (PNAS) Early Edition, the researchers used the computing power of Gordon and Comet, based at the San Diego Supercomputer Center (SDSC) at UC San Diego; and Stampede, at the Texas Advanced Computing Center at the University of Texas at Austin, to perform an unprecedented survey of protein structures using accelerated molecular dynamics or aMD – a method that performs a more complete sampling of the myriad shapes and conformations that a target protein molecule may go through.
The computing resources were provided by the National Science Foundation-funded Extreme Science and Engineering Discovery Environment (XSEDE) program, one of the most advanced collections of integrated digital resources and services in the world.
“The supercomputing power of Gordon, Comet, and Stampede allows us to run hundreds-of-nanosecond aMD simulations, which are able to capture millisecond timescale events in complex biomolecules,” said the study’s first author Yinglong Miao, a research specialist with the Howard Hughes Medical Institute at UC San Diego and research scientist with the UC San Diego Department of Pharmacology.
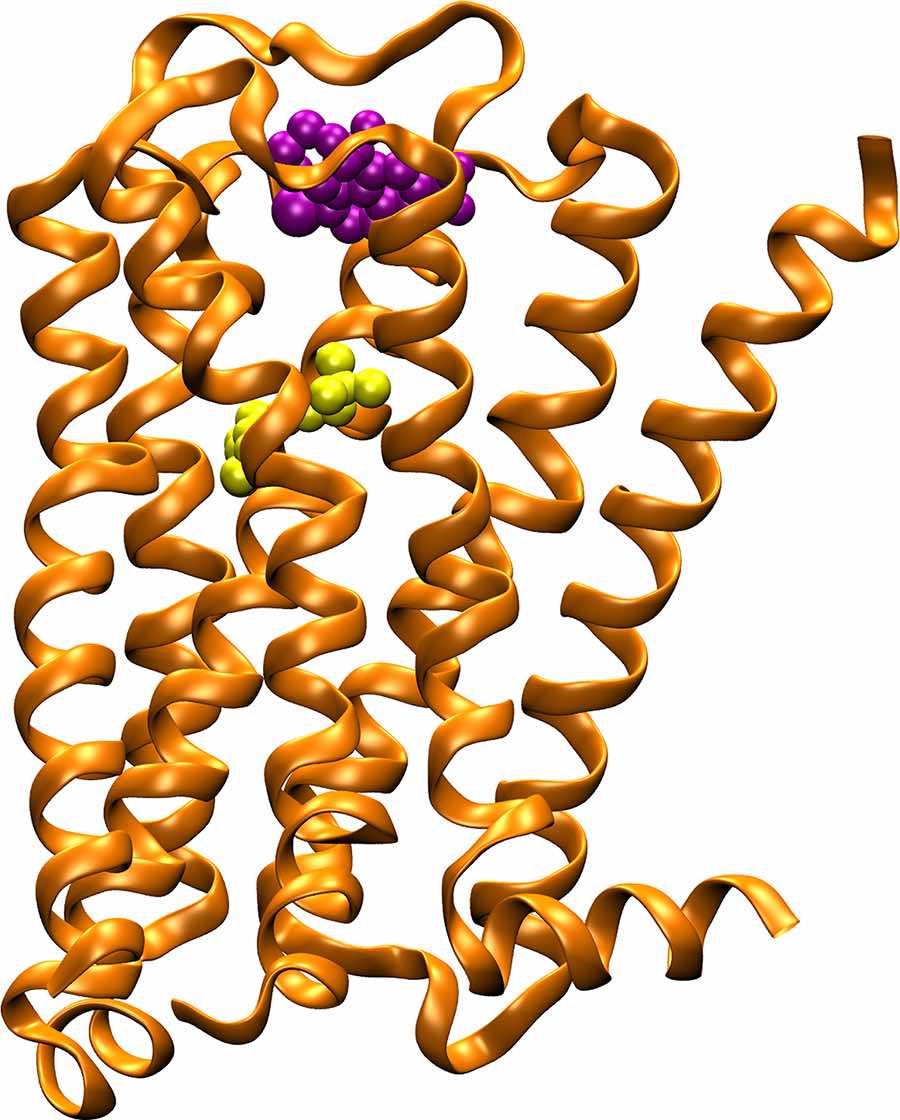
Though effective in most cases, today’s heart medications – many of which act on M2 muscarinic acetylcholine receptors or M2 mAChRs that decrease heart rate and reduce heart contractions – may carry side effects, sometimes serious. That’s because the genetic sequence of M2 mAChR’s primary ‘orthosteric’ binding site is “highly conserved,” and found in at least four other receptor types that are widely spread in the body, yielding unwanted results.
For this reason, drug designers are seeking a different approach, homing in on molecular targets or so-called “allosteric binding sites” that reside away from the receptor’s primary binding site and are built around a more diverse genetic sequence and structure than their counterpart ‘orthosteric’ binding sites. Essentially, allosteric modulators act as a kind of cellular dimmer-switch that, once turned on, ‘fine tunes’ the activation and pharmacological profile of the target receptor.
Allosteric sites typically exhibit great sequence diversity and therefore present exciting new targets for designing selective therapeutics,” said the study’s co-investigator J. Andrew McCammon, the Joseph E. Mayer Chair of Theoretical Chemistry, a Howard Hughes Medical Institute investigator, and Distinguished Professor of Pharmacology, all at UC San Diego. McCammon was recently named the winner of the 2016-17 Joseph O. Hirschfelder Prize in Theoretical Chemistry, awarded by the Theoretical Chemistry Institute at the University of Wisconsin-Madison.
In particular, drug designers have begun to aggressively search for allosteric modulators to fine-tune medications that bind to G protein-coupled receptors (GPCRs), the largest and most diverse group of membrane receptors in animals, plants, fungi and protozoa. These cell surface receptors act like an inbox for messages in the form of light energy, hormones and neurotransmitters, and perform an incredible array of functions in the human body.
In fact, between one-third to one-half of all marketed drugs act by binding to GPCRs, treating diseases including cancer, asthma, schizophrenia, Alzheimer’s and Parkinson’s disease, and heart disease.
More Targeted Therapies
Though many of the GPCR drugs have made their way to the medicine cabinet, most — including M2 mAChR targeted drugs — exhibit side effects owing to their lack of specificity. All these drugs target the orthosteric binding sites of receptors, thus creating the push to find more targeted therapies based on allosteric sites.
The problem here is that molecules that bind to these allosteric sites have proven extremely difficult to identify using conventional high-throughput screening techniques,” said McCammon, also a chemistry and biochemistry professor in UC San Diego’s Division of Physical Sciences.
Enter accelerated molecular dynamics and supercomputing. As described in this latest study, called Accelerated structure-based design of chemically diverse allosteric modulators of a muscarinic G protein-coupled receptor, some 38 lead compounds were selected from a database of compounds from the National Cancer Institute, using computationally enhanced simulations to account for binding strength and receptor flexibility. About half of these compounds exhibited the hallmarks of an allosteric behavior in subsequent in vitro experiments, with about a dozen showing strong affinity to the M2 mAChR binding site. Of these, the researchers highlighted two showing both strong affinity and high selectivity in studies of cellular behavior. These cutting-edge experiments were performed by collaborators from the Monash Institute of Pharmaceutical Sciences.
To our knowledge, this study demonstrates for the first time an unprecedented successful structure-based approach to identify chemically diverse and selective GPCR allosteric modulators with outstanding potential for further structure-activity relationship studies,” the researchers wrote.
The next steps will involve an investigation of the chemical properties of these novel molecules by the molecular chemists from Monash, led by Celine Valant and her colleague Arthur Christopoulos.
This is just the beginning. We believe that it will be possible to apply our combined cutting-edge in silico and in vitro techniques to a wide array of receptor targets that are involved in some of the most devastating diseases,” said Valant, the study’s co-lead investigator from Monash.
Also participating in the study were Dahlia Goldfeld, from the UC San Diego Department of Pharmacology; and Ee Von Moo and Patrick M. Sexton from Monash University.
Funding for this research was provided by grants from the National Science Foundation (MCB1020765, the National Institutes of Health (GM31749), Howard Hughes Medical Institute, the National Biomedical Computation Resource (NBCR), and the National Health and Medical Research Council (NHMRC) of Australia (APP1055134; APP1082318). In addition to XSEDE, supercomputing time was also provided by the Hopper and Edison supercomputers through the National Energy Research Scientific Computing Center (NERSC).