
Margo Brinton from Georgia State University
Researchers are using new techniques with HPC to learn more about how the West Nile virus replicates inside the brain.
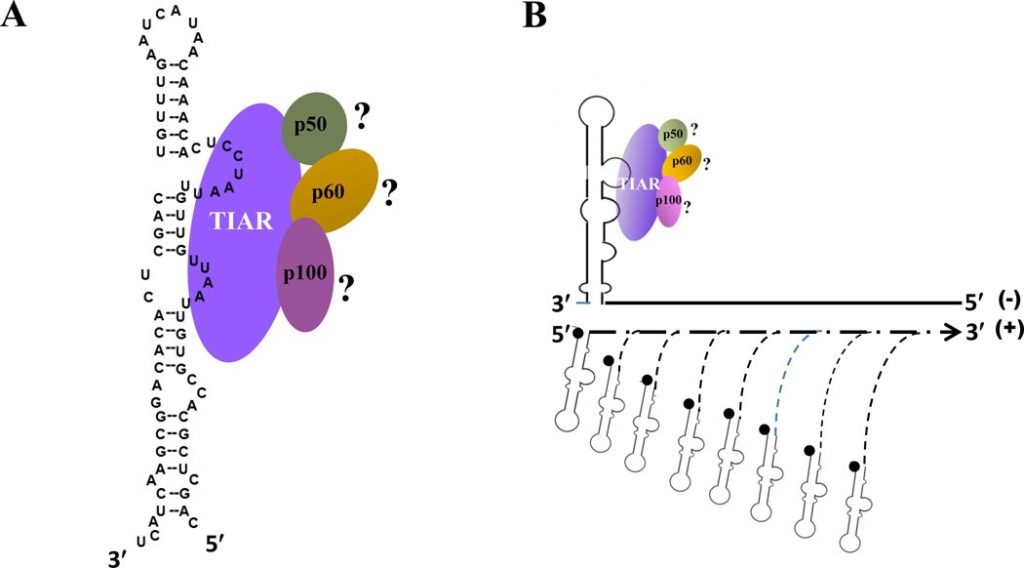
For most of her career, Margo Brinton at Georgia State University has worked on uncovering clues about how the West Nile virus uses cell proteins to facilitate its replication. The viral RNA genome is small, encoding only 10 proteins. In infected cells, cell proteins are commandeered by interaction with viral components to provide additional virus-specific functions.
Brinton, a Regents’ Professor in the Department of Biology, says the West Nile virus genome RNA is in general similar to a cell messenger RNA. However, in addition to being used to translate protein, the viral RNA also functions as a template for RNA replication. Conserved terminal viral stem loop RNA structures have been shown to be important in regulating the initiation of RNA synthesis and cell proteins are thought to interact with these viral stem loop RNA structures and function as “transcription factors” to facilitate viral RNA replication.
Margo Brinton, Regents’ Professor in the Department of Biology, Georgia State University In a paper published by Brinton and colleagues in Analytical Chemistry in January 2017, the researchers describe how they previously used the viral stem loop RNAs to pull out cell proteins that specifically interact with them. “That’s how we initially found TIAR, which is a cell protein that interacts with one of the viral stem loop RNAs,” Brinton said.
The result: the researchers found that the stoichiometry of the interaction was four TIAR proteins to one viral stem loop RNA molecule.
This finding told us that this viral RNA-cell protein interaction is more complex than we expected,” Brinton explained. “Based on previous data that was obtained under test tube conditions, we speculated that there were two protein binding sites on the viral stem loop RNA and finding four protein in the complex was surprising. Whether one RNA molecule binds to four individual TIAR molecules, or two dimers, or one tetramer, with the RNA functioning as an oligomerization enhancer, must next be determined.
Functions of the 3′ and 5′ genome RNA regions of members of the genus Flavivirus.
The research was enabled by a new technology that could have a broad impact by expanding the biological systems that scientists can study.
Borries Demeler, an author on the paper and a biophysicist at The University of Texas Health Science Center in San Antonio, develops software for analytical ultracentrifugation, a biophysical technique that spins biopolymers and protein molecules at high speed to characterize how they interact. This technology can separate biological macromolecules by spinning them up to 60,000 revolutions per minute, generating forces that are about 300,000 times as strong as the Earth’s gravity.
We can make molecules move inside a solution,” Demeler said. “Imagine that you have a protein solution and there’s an enzyme that you’re studying. You can make it sediment and diffuse inside of a chamber that you can monitor while the machine is spinning with the electronics that are built into the machine.”
“The nice thing about this technique is that the analysis is done in an environment that mimics what’s going on inside a real cell — it’s called a physiological environment,” Demeler said. “The fact that we can measure an interaction in a physiological environment similar to what is inside a cell and replicate what these molecules do when they come together is very important.”
Analytical ultracentrifugation allows one to study a molecule’s shape, size, and density as well as how proteins associate with other molecules and how strongly they bind to each other. “There are also many other questions we can answer with this technology,” Demeler said.
Borries Demeler, Biophysicist, The University of Texas Health Science Center in San Antonio With a new detector available now on a new instrument, the researchers can identify not only the size, shape and density, but also the material’s spectral properties, which suggest its composition and allow one to identify the type of complex formed. Demeler added that spectral properties of interacting molecules help researchers to discover new drug targets, and assist with the design of new drugs that can inhibit that interaction.
Theodor Svedberg, a Swedish chemist, invented the analytical ultracentrifuge in 1925, and was awarded the Nobel Prize in Chemistry in 1926 for his research on colloids and proteins using the analytical ultracentrifuge. Ever since, biochemists have used this technology to study biological macromolecules like proteins and DNA.
“It’s a very powerful method and a lot of researchers depend on it,” Demeler said.
Over several years, Demeler has developed analysis software for experiments performed with analytical ultracentrifuges. The goal is to facilitate the extraction of all of the information possible from the available data.
To do this, we developed very high-resolution analysis methods that require high performance computing (HPC) to access this information,” he said. “We rely on HPC. It’s absolutely critical.”
Most of the heavy lifting is done with UltraScan, an HPC enabled software developed by Demeler. As a researcher within the University of Texas System, Demeler has access to the Lonestar 5 supercomputer at the Texas Advanced Computing Center (TACC) at UT Austin.
Lonestar 5 is a very important resource,” he said. The supercomputer is provided through “The University of Texas System Research Cyberinfrastructure (UTRC)”. This initiative provides advanced computing, data storage, and high bandwidth data access among all 14 UT System institutions.
“The calculations that underlie the fitting of experimental data requires finite element solutions and esoteric optimization routines that require a lot of computational power.”
The UltraScan project also has a community allocation through the eXtreme Science and Engineering Discovery Environment (XSEDE) using Stampede2 (TACC), Jetstream (TACC), and Comet at the San Diego Supercomputer Center. “These are the main resources we’re using,” Demeler said.
The XSEDE community allocation offers the use of a network of software tools that were developed as a science gateway. This enables entire communities of users associated with a common scientific goal to use national resources through a common interface.
In addition to the software that resides on the HPC clusters, the team built a web-based gateway infrastructure that helps other researchers who may not be familiar with modelling analytical ultracentrifugation data. According to Demeler, many researchers have requested access to the UltraScan Science Gateway, and take advantage of this resource where they can upload their data and store it on a remotely hosted database.
Every institution has their own database where they host their data on the website, and we manage the website here at the UT Health Science Center in San Antonio,” Demeler said. “The website contains the programs needed for accessing the data, selecting the data, and then sending it off to the supercomputer of their choice.”

Borries Demeler, Biophysicist, The University of Texas Health Science Center in San Antonio
Importantly, these researchers get the benefit of doing top-notch science with high-resolution analysis by HPC without ever having to have expertise in biophysics or computer science. All of the analyses are done via web interface, which provides computing cycles — transparently to any user — through an NSF/XSEDE allocation grant awarded to Demeler.
A New Detector
In recent years, Demeler has begun using a new multi-wavelength detector that was developed for analytical ultracentrifugation — it’s more precise, but data analysis requires even more computing muscle.
“This is really exciting,” Demeler said. “The problem was already computationally demanding for single wavelength experiments and has now gotten that much bigger just by the introduction of the new detector. It turns out that you can beautifully parallelize these data because each wavelength can be analyzed independently, which has essentially multiplied the number of jobs that we’re submitting simultaneously.”
With access to computational resources, we are solving innovative problems and discovering revolutionary ways of looking at cellular systems and biological structure/function questions that can’t be solved any other way.”
Borries Demeler, The University of Texas Health Science Center in San Antonio
“That’s a big revolution,” he said. “It turns out the experiments that we can analyze now are much more informative. And we can ask very different questions that were previously essentially out of reach.” “Being able to understand on a molecular basis, the structure/function relationship for the interaction of a conserved viral stem loop RNA and a cell protein is an initial step in coming up with novel antiviral drug targets for West Nile virus infections.”
The results from this study contribute to the overall body of biological knowledge. But they also represent the first application of multi-wavelengths ultracentrifugation to a discrete protein/RNA interaction problem.
Demeler concluded: “With access to computational resources, we are solving innovative problems and discovering revolutionary ways of looking at cellular systems and biological structure/function questions that can’t be solved any other way.”
Source: TACC




