This feature continues our series of articles that survey the landscape of HPC and AI. This post focuses on use cases that show how AI technology — like an optimized technique of Diffusion Compartment Imaging (DCI) — is growing the potential of HPC.
Boston Children’s Hospital
Simon Warfield, Director of the Computational Radiology Laboratory at Boston Children’s Hospital and Professor of Radiology at Harvard Medical School, is working to use AI to better diagnose various forms of damage in the human brain including concussions, autism spectrum disorder and Pediatric Onset Multiple Sclerosis. The end goal will be a tool that allows clinicians to look directly at the brain rather than attempting to diagnose problems—and efficacy of treatment—by somewhat subjective assessments of cognitive behavior.
Collecting and Processing the Data
Professor Warfield and his team have created a Diffusion Compartment Imaging (DCI) technique, which is able to extract clinically relevant information about soft tissues in the brain. DCI is an improvement to Diffusion-Weighted Imaging (DWI) that works by using pulsing magnetic field gradients during an MRI scan to measure the atomic-scale movements of water molecules (called Brownian motion). The differential magnitude and direction of these movements provides the raw data needed to identify microstructures in the brain and to diagnose the integrity and health of neural tracts and other soft-tissue components.
[clickToTweet tweet=”A new optimized DCI technique can faster extract clinically relevant information faster about soft tissues in the brain. #AI” quote=”A new optimized DCI technique can faster extract clinically relevant information about soft tissues in the brain. #AI”]
However, there are several challenges to make DCI a clinically useful tool. For example, it can take about an hour to perform a single DCI scan when aiming at a very high spatial resolution, or about 10 minutes for most clinical applications. Before optimization, it took over 40 hours to process the tens of gigabytes of resulting water diffusion data.
Such long processing time made DCI completely unusable in many emergency situations, and further it made Diffusion Compartment Imaging challenging to fit into the workflow of today’s radiology departments. After optimization, it now takes about an hour to process the data.
Professor Warfield and Boston Children’s Hospital have been an Intel Parallel Computing Center (IPPC) for several years. This gave Professor Warfield’s team an extraordinary opportunity to optimize their code.
The results of the optimization work were transformative, providing a remarkable 75X performance improvement when running on Intel Xeon processors and a 161X improvement running on Intel Xeon Phi processors. A complete Diffusion Compartment Imaging study can now be completed in 16 minutes on a workstation, which means Diffusion Compartment Imaging can now be used in emergency situations, in a clinical setting, and to evaluate the efficacy of treatment. Even better, higher resolution images can be produced because the optimized code scales.
Volume Inferencing to Find Brain Damage
Data processing isn’t the only challenge with using Diffusion Compartment Imaging in clinical settings as a scan typically generates tens to hundreds of images. Many radiologists are finding it difficult to keep up with the increasing volume of work.
Professor Warfield believes AI is a solution to this problem. The vision is to train a model that can automatically and quickly sort through hundreds of images to pinpoint those that differ from comparable images of a healthy brain. The goal is to provide the radiologist with the essential highlights of a complex study, identifying not only the most relevant images, but also pinpointing the critical areas on those images.
Such pinpoint inferencing can be seen in the next case study.
Detecting Cancer Cells More Efficiently
Cancer cells are to be detected and classified more efficiently and accurately, using groundbreaking artificial intelligence—thanks to a new collaboration between the University of Warwick, Intel Corporation, the Alan Turing Institute and University Hospitals Coventry & Warwickshire NHS Trust (UHCW). The collaboration is part of The Alan Turing Institute’s strategic partnership with Intel.
We have long known that important aspects of cellular pathology can be done faster with computers than by humans. — Professor David Snead, clinical lead for cellular pathology and director of the UHCW Centre of Excellence.
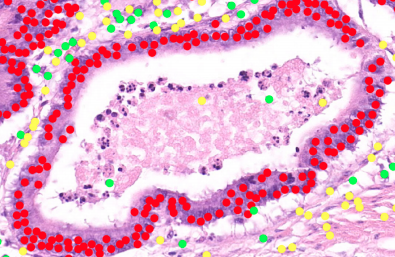
Scientists at the University of Warwick’s Tissue Image Analytics (TIA) Laboratory—led by Professor Nasir Rajpoot from the Department of Computer Science—are creating a large, digital repository of a variety of tumour and immune cells found in thousands of human tissue samples, and are developing algorithms to recognize these cells automatically.
Cancer tumor cells are highlighted in red
(Source: University of Warwick)
Professor Rajpoot commented: “The collaboration will enable us to benefit from world-class computer science expertise at Intel with the aim of optimizing our digital pathology image analysis software pipeline and deploying some of the latest cutting-edge technologies developed in our lab for computer-assisted diagnosis and grading of cancer.”
The digital pathology imaging solution aims to enable pathologists to increase their accuracy and reliability in analyzing cancerous tissue specimens over what can be achieved with existing methods.
“We have long known that important aspects of cellular pathology can be done faster with computers than by humans,” said Professor David Snead, clinical lead for cellular pathology and director of the UHCW Centre of Excellence.
The insideHPC Special Report on AI-HPC will also cover the following topics over the next few weeks:
- An Overview of AI in the HPC Landscape
- AI and HPC: Inferencing, Platforms & InfrastructureAdditional Customer Use Cases
- The AI Software Ecosystem
- Hardware to Support the AI Software Ecosystem
Download the full report: “insideHPC Special Report: AI-HPC is Happening Now” courtesy of Intel.