Reanalysis of public genetic data using innovative computational methods has allowed the identification of new genetic markers associated with an increased risk to develop Type 2 Diabetes (T2D).
The study, led by Barcelona Supercomputing Center and published today in Nature Communications, represents a new way of exploiting preexisting genetic data to obtain new and relevant discoveries for genetics and biomedicine, highlighting the importance of data sharing initiatives and policies in science.
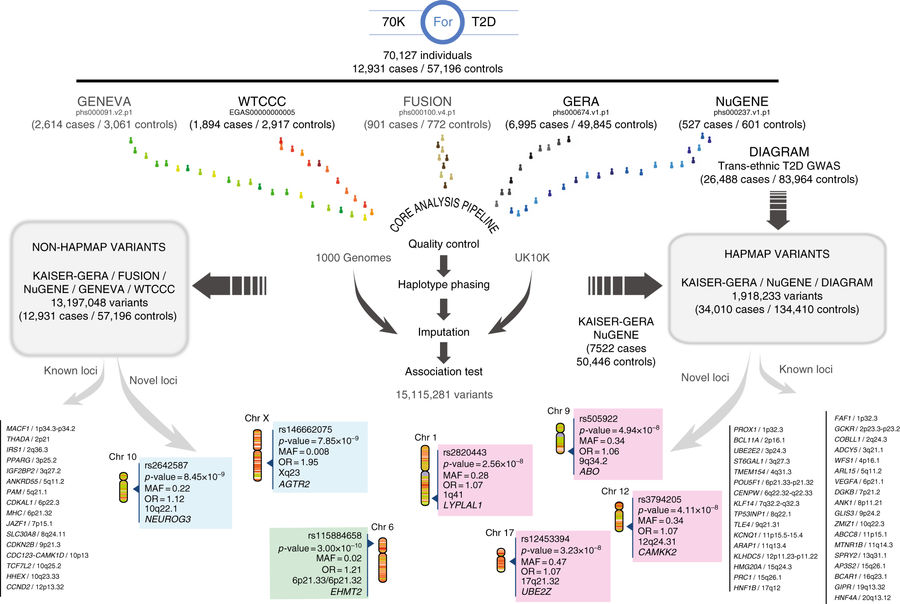
Discovery and replication strategy. Publicly available GWAS datasets representing a total of 12,931 cases and 57,196 controls (70KforT2D) were first quality controlled, phased, and imputed, using 1000G and UK10K separately.
Due to its physiological and genetic complexity, current treatment and prevention protocols for T2D are not yet efficient. Understanding its underlying genetic basis is key to allow the identification of new targets for therapies, as well as the design of efficient prevention strategies.
This study has generated and applied innovative approaches for GWAS analysis to publicly available genetic data for around 70.000 individuals, reaching unprecedented levels of genetic resolution. This method led to the identification of seven novel regions that are associated with increased risk for type 2 diabetes. Although around 100 regions of the genome have been reported to be associated with increased T2D susceptibility, most of them modify the risk in the range of 5 to 30%. One of the novel regions reported in this study corresponds to a rare genetic marker in the X chromosome that increases the risk for T2D by 200% in males. This suggests that the identification of such marker prior the development of T2D could be useful to design and apply preventive strategies that delay or avoid the development of this disease. Furthermore, this study shed light on the potential mechanism behind this increased risk, pointing the gene AGTR2, as a potential target to develop efficient treatments.
By re-analyzing public genetic data in-house, we had the opportunity to deeply explore the role of genetic variation from the X chromosome, which is often neglected from genetic analysis. Our findings underscore that novel insights into disease biology have been missed for the mere fact of excluding this data” says Sílvia Bonàs-Guarch, first author of the study.
David Torrents, ICREA Research Professor and Computational Genomics group manager at BSC and Josep M. Mercader, collaborator at the same institution, have supervised the project. In addition, internal BSC efforts also included the activity of the Workflows and Distributed Computing group led by Rosa M. Badia.
The sharing of data in biomedicine allows its reanalysis using newer and more efficient approaches and to answer more ambitious questions regarding the basis of disease, as we have done in this study for type 2 diabetes”, underlines David Torrents.
The continuous increase of the production of data in biomedicine, mostly due to the advances in DNA and RNA sequencing technologies, allows the search of the genetic and molecular basis of disease at an unprecedented speed and resolution. Normally, in a typical genetic study, the generation (sequencing or genotyping) of thousands of genetic profiles from patients of a particular disease is followed by complex analyses that result in findings that can ultimately inform the diagnosis, prognosis or treatment markers for a particular disease. These data is then stored in large databases, for example the European Genome and Phenome Archive (EGA), where is subjected to different sharing policies. Whereas the scientific community is globally promoting open access policies, respecting patient’s privacy and rights, some sectors choose to keep this data private, usually to avoid concurrence at commercial and at scientific level.
We honestly were surprised by the amount of additional information that can be obtained by reanalyzing the same data with novel computational and genetic resources”, says Josep M. Mercader, co-supervisor of this work.



