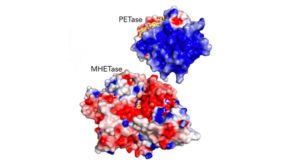
The dynamic enzyme duo of PETase and MHETase break down PET plastic, commonly found in disposable bottles.
Plastic waste is a big problem in the environment. About 300 million tons is produced every year, according to the United Nations. Much of that is polyethylene terephthalate (PET), used to make single-use plastic bottles, carpets, and clamshell packaging. In the U.S., the Environmental Protection Agency estimates annually that only about 29 percent of PET bottles are recycled.
About 300 million tons of global plastic waste is produced each year. Much of that is polyethylene terephthalate (PET) plastic, used to make single-use beverage bottles like the ones shown. In 2016, Japanese scientists discovered bacteria with an enzyme called PETase that degrades PET. In 2020, scientists used supercomputers to resolve the structure of a partner enzyme, called MHETase, that helps PETase eat plastic.
In 2016, Japanese scientists discovered that the bacteria Ideonella sakaiensis had evolved digestive enzymes called PETase that breakdown PET. And in 2020, supercomputers have helped reveal more about a sidekick enzyme, called MHETase, that helps PETase breakdown PET plastic. While dealing with plastic pollution at scale remains daunting, in the words of Jeff Goldblum’s character in the movie Jurassic Park, “Life finds a way.”
“This most recent study focuses on the partner enzyme of PETase, which is called MHETase. It works on mono(2-hydroxyethyl) terephthalate (MHET), one of the plastic breakdown molecules that PETase produces,” said Brandon Knott, a staff engineer at the National Renewable Energy Lab (NREL) located in Golden, Colorado. Knott co-authored a study on MHETase published in the Proceedings of the National Academy of Sciences.
The work presented in the study includes crystal structures obtained of MHETase and how those compared to PETase.
“There are some aspects in which they are quite similar, including their catalytic residues,” Knott explained. “But there are other aspects where they’re quite different, which leads to them having different substrate specificities.”
These differences help understand why they work well together, where MHETase cleans up what PETase can’t.

NREL scientists and co-authors of MHETase PNAS study: Erika Erickson and Brandon Knott credit: NREL
“Very likely, these two enzymes were co-opted towards this ability to degrade plastic from another function that they had naturally, whether that’s to break down biomass of some sort; or another way to gain a sugar source or a nutrient source in the environment,” said co-author Erika Erickson, a postdoctoral researcher at NREL.
Erickson led the wet lab investigation of MHETase, expressing and purifying it to characterize what substrates it’s able to break down, how quickly, and what are the limitations of that reaction. She also explored different MHETase mutations to see if they improved or abolished activity and help verify that the active site is indeed the predicted residues.
“There’s a synergistic behavior between PETase and MHETase,” Erickson explained. “While PETase is very exciting on its own, when friends are involved it’s a much more efficient process. If you think about the PETase enzyme interacting with the surface of a polymer film or some other solid substrate, and it’s releasing small chunks into the aqueous phase, then you might have this second enzyme that’s there to catch those small chunks to continue breaking them down.”
Knott added that pivotal to the paper was quantifying how much MHETase helps PETase in terms of how much PET plastic is broken down.
“By adding MHETase into the mix with PETase, PET is broken down twice as well,” he said. “Add the chimera enzyme where they’re called covalently bonded together, and you get another factor of three improvement in the PET depolymerization. Altogether going from PETase alone to the chimera enzyme, you have a six-fold improvement in terms of PET depolymerization.”
 The team did quite a bit of biology and biochemistry to look at how the two enzymes work separately and how they work when they’re together. They used bioinformatics to look at possible evolution of the MHETase enzyme. They also employed molecular dynamics simulations to elucidate the MHETase reaction mechanism. And they studied the effects of covalently binding the two partner enzymes together.
The team did quite a bit of biology and biochemistry to look at how the two enzymes work separately and how they work when they’re together. They used bioinformatics to look at possible evolution of the MHETase enzyme. They also employed molecular dynamics simulations to elucidate the MHETase reaction mechanism. And they studied the effects of covalently binding the two partner enzymes together.
Knott’s team was awarded supercomputer allocations to run molecular dynamics simulations of MHETase and PETase in their plastic degradation research by XSEDE, the Extreme Science and Engineering Discovery Environment, funded by the National Science Foundation. XSEDE-allocated supercomputers Stampede2 at the Texas Advanced Computing Center and Comet at the San Diego Supercomputer Center were used for this study. The NREL Eagle supercomputer supported by the DOE Office of Energy Efficiency and Renewable Energy was also used in the study.
 “Our group has been convinced for a very long time that coupling computer simulations with wet lab experiments, with crystal structures, and with various other techniques is really powerful,” Knott said. The simulations allowed the team to add a dynamic complement to the static crystal structures that otherwise would not have been feasible in their investigation of the reaction mechanisms.
“Our group has been convinced for a very long time that coupling computer simulations with wet lab experiments, with crystal structures, and with various other techniques is really powerful,” Knott said. The simulations allowed the team to add a dynamic complement to the static crystal structures that otherwise would not have been feasible in their investigation of the reaction mechanisms.
Corresponding author Gregg Beckham of NREL has employed XSEDE resources continuously since 2009 in the context of investigating biological systems to degrade recalcitrant natural polymers such as cellulose, and now in this case PET plastic. In 2018, Beckham’s group used XSEDE to solve the structure of PETase.
In this latest study they used simulation to find the rate limiting step of reaction, whether the ethylene glycol released by the PETase hangs around the active site of MHETase and helps in the reaction, or does it leave the scene?
The simulations showed that ethylene glycol leaves the active site between step one and step two after it’s cleaved off. Then step two proceeds, absent of that ethylene glycol molecule.
“Those kinds of questions we can get at in detail with pretty exquisite time and space resolution with the molecular simulations. They would be very difficult or impossible to answer with experiment, given the space and time resolution that we have available to us in molecular simulation,” Knott said.
“The XSEDE resources are critical in helping us to overcome some of the challenges and to really facilitate making that work possible,” he added.
For software, Knott’s team used CHARMM for building enzyme and polymer systems. The longer simulation runs in the study used the molecular dynamics software NAMD. They used Amber for the QM/MM reaction mechanism studies, where the active site was treated with a semi-empirical quantum mechanical level of theory. The rest of the enzyme and solvation were treated with classical mechanics. “We’re using several different pieces of software on those two machines,” Knott said.
“Having these reliable resources from XSEDE with such a high capacity, in terms of the number of high-performance nodes, professionally administered, and the high level of support is essential to the science that we’ve been able to accomplish on this front,” Knott added.
While still far out from a solution that deals with the scale of the global plastic waste problem, both scientists see reason for hope.
“Some companies have started to truly make progress in this direction, that there really could be some enzymatic or biological processes for degrading certain types of plastic waste. But there’s still a lot of work to do here in optimization and achieving a scale that will make a real difference, and not just be a demonstration,” Erickson said.
“One of the advantages, you have of doing this biologically is that the breakdown to monomers gives you a lot of flexibility, not only to recycle plastic and displace that petroleum usage and divert waste from landfills or the environment, but also the flexibility to produce virgin quality PET again,” Knott added.
Both scientists also agreed that it’s remarkable that nature rapidly identified a possible strategy to help with humanity’s plastic waste problem.
Said Erickson: “This isn’t going to be a magic bullet that fixes our situation and saves us from ourselves. This may be one of many solutions. And I think the impact of this type of research is for creativity in the minds of the public to remind them that this is an issue. There’s still a lot of work to do.”
The study, “Characterization and engineering of a two-enzyme system for plastics depolymerization,” was published in October 2020 in the Proceedings of the National Academy of Sciences. The authors are Brandon C. Knott, Erika Erickson, Japheth E. Gado, Isabel Pardo, Ece Topuzlu, Jared J. Anderson, Graham Dominick, Christopher W. Johnson, Nicholas A. Rorrer, Caralyn J. Szostkiewicz, and Gregg T. Beckham of the Renewable Resources and Enabling Sciences Center, National Renewable Energy Laboratory; Mark D. Allen, Rosie Graham, Harry P. Austin, and John E. McGeehan of the Centre for Enzyme Innovation, School of Biological Sciences, Institute of Biological and Biomedical Sciences, University of Portsmouth; Fiona L. Kearns and Lee Woodcock of the Department of Chemistry, University of South Florida; Valérie Copié of the Department of Chemistry and Biochemistry, Montana State University; Christina M. Payne of the Department of Chemical and Materials Engineering, University of Kentucky, and Bryon S. Donohoe of the Biosciences Center, National Renewable Energy Laboratory. Funding was provided by the DOE, Office of Energy Efficiency and Renewable Energy, Advanced Manufacturing Office and Bioenergy Technologies Office. Computer time was provided by Extreme Science and Engineering Discovery Environment allocation MCB-090159 at the San Diego Supercomputer Center and the Texas Advanced Computing Center, and by the National Renewable Energy Laboratory Computational Sciences Center supported by the DOE Office of Energy Efficiency and Renewable Energy under Contract DE-AC36-08GO28308.
source: Jorge Salazar, science writer, Texas Advanced Computing Center




