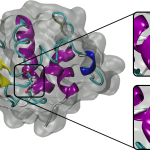
Over the past decade, teams of engineers, chemists and biologists have analyzed the physical and chemical properties of cicada wings, hoping to unlock the secret of their ability to kill microbes on contact. If this function of nature can be replicated by science, it may lead to products with inherently antibacterial surfaces that are more […]
OLCF and Stony Brook: Using HPC Simulations to Learn How Cicada Wings Kill Bacteria
EXAALT-ing Molecular Dynamics to the Power of Exascale
As part of the Exascale Computing Project (ECP), a collaborative team of scientists, software developers, and hardware integration specialists from across the Department of Energy (DOE) has developed the Exascale Atomistics for Accuracy, Length, and Time (EXAALT) application to bring molecular dynamics (MD) into the exascale era. Danny Perez, a physicist within the Theoretical Division at Los Alamos National Laboratory and the project’s principal investigator says, “We’ve implemented new scalable methods that allow us to access as much of the accuracy, length and time space as possible on exascale machines by rethinking our methods and developing algorithms that go around some of the bottlenecks that limited scaling previously.” Such a capability has potential to revolutionize MD.
Compendium of articles published on Numerical Algorithms for HPC Science
The Royal Society Publishing has recently released a special compendium of articles based on a recent scientific discussion meeting with HPC Industry thought leaders. “This issue contains contributions from those who develop and implement numerical algorithms and software libraries – numerical analysts, computer scientists, and high-performance computing researchers – with those who use them in some of today’s most challenging applications.”
Podcast: ECP EXAALT Program Extends the Reach of Molecular Dynamics
Computationally, EXAALT’s goal is to develop a comprehensive molecular dynamics capability for exascale. “The user should be able to say, ‘I’m interested in this kind of system size, timescale, and accuracy,’ and directly access the regime without being constrained by the usual scaling paths of current codes,” said Danny Perez of Los Alamos National Laboratory (LANL) and the EXAALT team.
BioScience gets a Boost Through Order-of-Magnitude Computing Gains
Intel is working with leaders in the field to eliminate today’s data processing bottlenecks. In this guest post from Intel, the company explores how BioScience is getting a leg up from order-of-magnitude computing progress. “Intel’s framework is designed to make HPC simpler, more affordable, and more powerful.”
Amber Molecular Dynamics Optimization on Intel Xeon Phi
“The Amber Molecular Dynamics software is a well known and understood application for the structural dynamics of large biological molecules. With modern computer systems, a speedup in the computations can lead to studying events that occur on longer timescales, as well as statistical convergences. By incorporating the Intel Xeon Phi coprocessor into the workflow on a compute server, additional speedups can be obtained.”
Gather Scatter Operations
Gather and scatter operations are used in many domains. However, to use these types of functions on an SIMD architecture creates some programming challenges.
Video: GPU-Accelerated Analysis of Large Biomolecular Complexes
“This presentation will highlight the use of GPU ray tracing for visualizing the process of photosynthesis, and GPU accelerated analysis of results of hybrid structure determination methods that combine data from cryo-electron microscopy and X-ray crystallography atom molecular dynamics with all- simulations.”