 Rice University synthetic biologists have won a $1.5 million grant from the National Science Foundation (NSF) to modify living cells to act as memory-storage devices, the university announced today.
Rice University synthetic biologists have won a $1.5 million grant from the National Science Foundation (NSF) to modify living cells to act as memory-storage devices, the university announced today.
NSF issued the award for research by principal investigator Jonathan Silberg, the Stewart Memorial Professor of Biochemistry, and his colleagues to make the equivalent of read-write-erase memories found in computers and other devices. Rice said these programmable cells, starting with E. coli, will capture and store information about their environments for later analysis by researchers.
“Cells read out dynamic information about their local habitat, process that information to make behavioral decisions and store some of that information in their biomolecules,” said Silberg, director of Rice’s Systems, Synthetic and Physical Biology Ph.D. program. “While cells have the potential to be integrated with semiconductor materials to create a whole new generation of sustainable technologies that read out information stored in biomolecules, we remain limited in our ability to interface cells and materials to create useful bioelectronic devices.”
The work at Rice recalls DNA-based data storage R&D at companies such as Catalog, whose CTO is former IBM HPC leader David Turek.
Rice co-principal investigators are Rafael Verduzco, a professor of chemical and biomolecular engineering and materials science and nanoengineering, James Chappell, an assistant professor of biosciences and bioengineering, and Caroline Ajo-Franklin, a professor of biosciences and bioengineering.
“Cells can already be programmed to act as read-only memory (ROM), storing their data in DNA,” Chappell said. “In fact, biological memory can achieve information densities that exceed commercial memory storage devices. However, mimicking random-access memory (RAM) and its multiple read-write-erase capabilities is the challenge we aim to solve.”
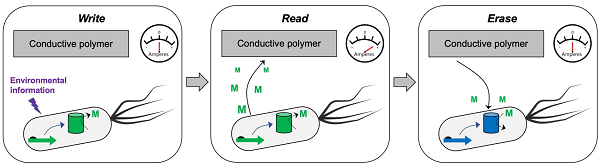
First, rather than encode data directly into DNA, the team will code hundreds of bytes of information within synthetic RNA biomolecules that reside within cells, which will also be programmed to synthesize redox-active small molecules that carry information to and from the outside world.
On the outside, the memory encoded within these redox-active molecules can be quickly read by semiconducting polymers, equivalent to the wires and associated electronics that carry information from a computer processor to a screen.
These will require an interface to read and write data. The researchers are designing printable hydrogel-based chambers that contain the microbes and associated electronics to read the small molecules that pass through the cells’ membranes.
This will allow the cells to be quickly programmed to capture information about a variety of targets. And all of this has to be done without harming the cells themselves.
The RAM created in these cells is expected to be stable for days to weeks or even longer, although that aspect of the proposed bioelectronic devices will require characterization to fully understand the technology’s potential, Silberg said.
He noted that all of this work will require expertise in many disciplines to create electrical interfaces between microbes and materials, including RNA biology, microbial electrochemistry, biomolecular engineering, metabolic engineering and semiconducting polymers.
“At Rice, we have diverse researchers in materials science and synthetic biology who are interested in interfacing advanced materials with microbial systems,” Silberg said.
source: Rice University



