 Calling it the largest federated learning study in the medical field to date, Intel Labs and Penn Medicine have announced a joint research study to help international healthcare and research institutions identify malignant brain tumors. The study, which involved a global dataset from 71 institutions across six continents, improved brain tumor detection, the two organizations said.
Calling it the largest federated learning study in the medical field to date, Intel Labs and Penn Medicine have announced a joint research study to help international healthcare and research institutions identify malignant brain tumors. The study, which involved a global dataset from 71 institutions across six continents, improved brain tumor detection, the two organizations said.
Data accessibility has long been an issue in healthcare because of state and national data privacy laws, such as the Health Insurance Portability and Accountability Act (HIPAA). Medical research and data sharing at scale have been almost impossible to achieve without compromising patient health information. But Intel said its federated learning – a distributed machine learning approach – hardware and software comply with data privacy concerns and preserve data integrity, privacy and security through confidential computing.
The Penn Medicine-Intel result was accomplished by processing high volumes of data in a decentralized system using Intel federated learning technology paired with Intel Software Guard Extensions (SGX), which removes data-sharing barriers that have historically prevented collaboration on similar cancer and disease research. Intel said the system addresses data privacy concerns by keeping raw data inside the data holders’ compute infrastructure and only allowing model updates computed from that data to be sent to a central server or aggregator, not the data itself.
The results of the Penn Medicine-Intel Labs research were published in the peer-reviewed journal, Nature Communications.
Said senior author Spyridon Bakas, PhD, assistant professor of Pathology & Laboratory Medicine and Radiology at Penn Medicine (formally called the Perelman School of Medicine at the University of Pennsylvania), “In this study, federated learning shows its potential as a paradigm shift in securing multi-institutional collaborations by enabling access to the largest and most diverse data set of glioblastoma patients ever considered in the literature, while all data are retained within each institution at all times. The more data we can feed into machine learning models, the more accurate they become, which in turn can improve our ability to understand and treat even rare diseases, such as glioblastoma.”
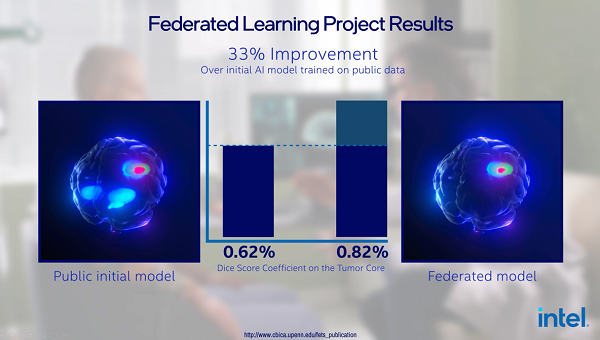 To advance the treatment of diseases, researchers must access large amounts of medical data – in most cases, datasets that exceed the threshold that one facility can produce. The research demonstrates the effectiveness of federated learning at scale and the potential benefits the healthcare industry can realize when multisite data silos are unlocked. Benefits include early detection of disease, which could improve quality of life or increase a patient’s lifespan.
To advance the treatment of diseases, researchers must access large amounts of medical data – in most cases, datasets that exceed the threshold that one facility can produce. The research demonstrates the effectiveness of federated learning at scale and the potential benefits the healthcare industry can realize when multisite data silos are unlocked. Benefits include early detection of disease, which could improve quality of life or increase a patient’s lifespan.
“Federated learning has tremendous potential across numerous domains, particularly within healthcare, as shown by our research with Penn Medicine,” said Jason Martin, principal engineer, Intel Labs. “Its ability to protect sensitive information and data opens the door for future studies and collaboration, especially in cases where datasets would otherwise be inaccessible. Our work with Penn Medicine has the potential to positively impact patients across the globe and we look forward to continuing to explore the promise of federated learning.”
In 2020, Intel and Penn Medicine announced the agreement to cooperate and use federated learning to improve tumor detection and improve treatment outcomes of a rare form of cancer called glioblastoma (GBM), the most common and fatal adult brain tumor with a median survival of just 14 months after standard treatment. While treatment options have expanded over the past 20 years, there has not been an improvement in overall survival rates. The research was funded by the Informatics Technology for Cancer Research program out of the National Cancer Institute of the National Institutes of Health.
 Penn Medicine and 71 international healthcare/research institutions used Intel’s federated learning hardware and software to improve the detection of rare cancer boundaries. A new state-of-the-art AI software platform called Federated Tumor Segmentation (FeTS) was used by radiologists to determine the boundary of a tumor and improve the identification of the “operable region” of tumors or “tumor core.” Radiologists annotated their data and used open federated learning (OpenFL), an open source framework for training machine learning algorithms, to run the federated training. The platform was trained on 3.7 million images from 6,314 GBM patients across six continents, the largest brain tumor dataset to date.
Penn Medicine and 71 international healthcare/research institutions used Intel’s federated learning hardware and software to improve the detection of rare cancer boundaries. A new state-of-the-art AI software platform called Federated Tumor Segmentation (FeTS) was used by radiologists to determine the boundary of a tumor and improve the identification of the “operable region” of tumors or “tumor core.” Radiologists annotated their data and used open federated learning (OpenFL), an open source framework for training machine learning algorithms, to run the federated training. The platform was trained on 3.7 million images from 6,314 GBM patients across six continents, the largest brain tumor dataset to date.
Through this project, Intel Labs and Penn Medicine have created a proof of concept for using federated learning to gain knowledge from data. The solution can significantly affect healthcare and other study areas, particularly among other types of cancer research. Specifically, Intel developed the OpenFL open source project to enable customers to adopt real-world cross-silo federated learning and confidently deploy it on Intel SGX. In addition, the novel FeTS initiative was established as a collaborative network to provide a platform for ongoing development and to encourage collaboration with the FeTS platform and Intel’s OpenFL open source toolkit, both available on GitHub.
source: Intel




