SuperMUC Cluster at LRZ in Germany
Researchers using the SuperMUC cluster in Germany have discovered a set of unknown species in rainforest soils. As described in a new paper published in Nature Ecology and Evolution, their study on microbial diversity in tropical rainforests required over one million CPU hours to complete.
Tropical rainforests are one of the most species-rich areas on earth. Thousands of animal and plant species live there. The smaller microbial protists, which are not visible to the naked eye, are also native to these forests, where they live in the soils and elsewhere. An international team of scientists, among them three HITS researchers, examined them more closely by analyzing their DNA. They discovered many unknown species, including many parasites, which may contribute to the stability of rainforest ecosystems.
“Without the outstanding high performance computing infrastructure in Germany and especially at LRZ, this study would not have been feasible. The availability of SuperMUC constitutes an essential national advantage in the international scientific competition,” said Alexandros Stamatakis, Heidelberg Institute for Theoretical Studies.
An international team of scientists led by Micah Dunthorn (Technische Universität Kaiserslautern, funded by the Emmy-Noether Program) had collected soil samples, including the microbes living in them, in lowland rainforests of Costa Rica, Panama, and Ecuador. Thereafter, they extracted and sequenced their DNA. Alexandros Stamatakis, Lucas Czech, and Alexey Kozlov (Heidelberger Institut für Theoretische Studien und Karlsruher Institut für Technologie) then analyzed the more than 130 million sequences using the SuperMUC supercomputer.
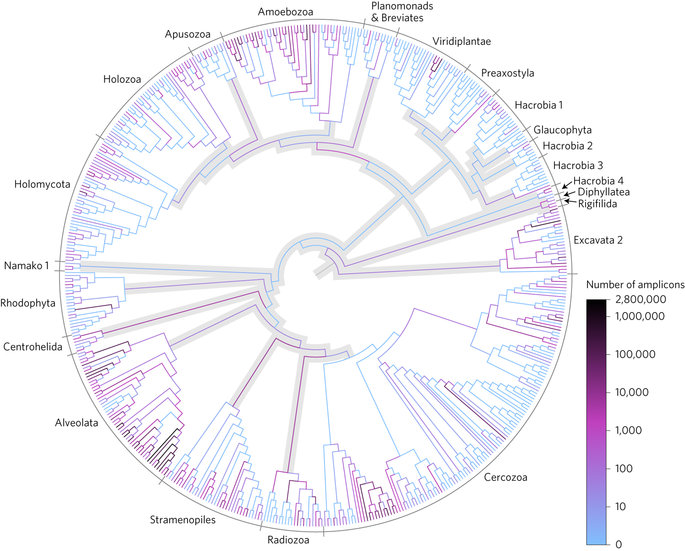
Phylogenetic placement of Neotropical soil protist reads on a taxonomically unconstrained global eukaryotic tree.
In 2014, Stamatakis had already contributed to a study with the computation of a phylogenetic tree of insects, featured on the cover page of Science. Later on, he had studied the phylogenetic tree of birds leveraging SuperMUC as well.
Over the years, Stamatakis‘ team was able to improve the algorithms and speed up the computations so that they now were able to perform this investigation of microbes in rainforests. Currently, the team works on a new approach to parallelize their software so that it will be able to handle datasets that are ten times as large.