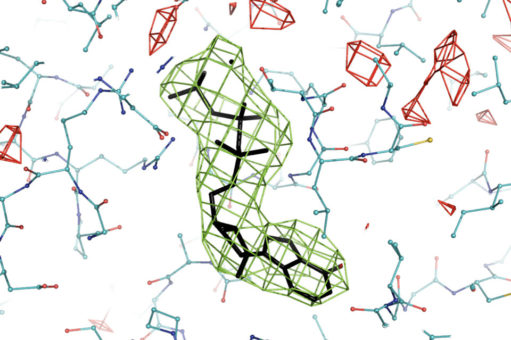
Pavel Afonine in Berkeley Lab’s Molecular Physics and Integrated Bioimaging Division created a PHENIX tool to calculate difference maps in real space from cryo-EM data. This figure shows such a map for one adenosine triphosphate molecule in a complex called the 26S proteasome. (Credit: Pavel Afonine/Berkeley Lab)
The National Institutes of Health (NIH) has awarded $9.3 million to the Department of Energy’s Lawrence Berkeley National Laboratory (Berkeley Lab) to support ongoing development of PHENIX, a software suite for solving three-dimensional macromolecular structures.
PHENIX, which stands for Python-based Hierarchical ENvironment for Integrated Xtallography, uses modern programming concepts and advanced algorithms to automate the analysis of structural biology data. The grant is awarded through the National Institute of General Medical Sciences at NIH.
Officially launched in 2000 with NIH funding, the project is a collaboration among researchers based at Berkeley Lab, Los Alamos National Laboratory, Cambridge University, and Duke University.
The impetus behind PHENIX is a desire to make the computational aspects of crystallography more automated, reducing human error and speeding solutions,” said PHENIX principal investigator Paul Adams, director of Berkeley Lab’s Molecular Biophysics and Integrated Bioimaging Division.
Knowing the precise location of each atom in a molecule in three dimensions is critical to understanding how the molecule functions and interacts with other molecules. The shapes of the proteins in a cell, for example, reveal a lot about the work they do—whether it’s providing structural support or motility, forming selectively permeable membrane channels, or powering the cell’s metabolism. DNA sequence alone still doesn’t give enough information about how a molecule folds, or how it changes conformation.
The software is openly and freely available for academic users. Over the years, the international community of crystallographers has contributed to PHENIX’s development through the open source Computational Crystallography Toolbox.
In the past 15 years, Adams noted, “the level of automation and computation algorithms that we’ve developed has really changed the way people do a lot of structural biology work.”
Recent improvements to PHENIX in the area of experimental phasing have enabled researchers to make use of noisier, lower resolution data that previously would have been discarded.
One major development in the field of structural biology over the past five years has been the ascendance of cryo-electron microscopy (cryo-EM), where a molecule is rapidly cooled and inserted into an electron microscope. The technology owes its current popularity to advances in electron detector technology, including a direct electron detector developed by Peter Denes and colleagues at the National Center for Electron Microscopy at Berkeley Lab’s Molecular Foundry, a DOE Office of Science User Facility.
Even before this revolution, as they’re calling it, took place we thought that cryo-EM would be an important area for us to get involved in,” Adams said.
He added that half of the work proposed under the new grant will be developing new methods for building, refining, and validating models in cryo-EM. The other half, he said, will be continuing the themes of extending crystallographic methods to work at lower resolution and with weaker data, and making it possible to solve molecular structures when there’s no similar model in the database.
This new cryo-EM approach can produce pictures that are nearly as clear as those from X-ray diffraction, but often with less work,” said Tom Terwilliger, whose group in the Bioscience Division at Los Alamos National Laboratory is a partner in the collaborative PHENIX project. “Our new tools will make it easier for researchers to interpret their pictures from cryo-EM, hopefully making this new technique as routine as X-ray crystallography is today.”
Adams noted that this kind of multi-institutional, multi-disciplinary, technology development is something at which the national labs excel.
We are very grateful to NIH for recognizing the value of the project and graciously agreeing to continue funding it,” he said. “Their commitment has been long-term; it’s been very impressive and has had a big impact on the research community.”
Other Berkeley Lab Biosciences MBIB collaborators on PHENIX include: Pavel Afonine, Dorothee Liebschner, Nigel Moriarty, Billy Poon, and Oleg Sobolev.