Technical Introduction
- GASNet-EX low-level layer that provides a network-independent interface suitable for Partitioned Global Address Space (PGAS) runtime developers
- UPC++ C++ PGAS library for application, framework and library developers, a productivity layer over GASNet-EX
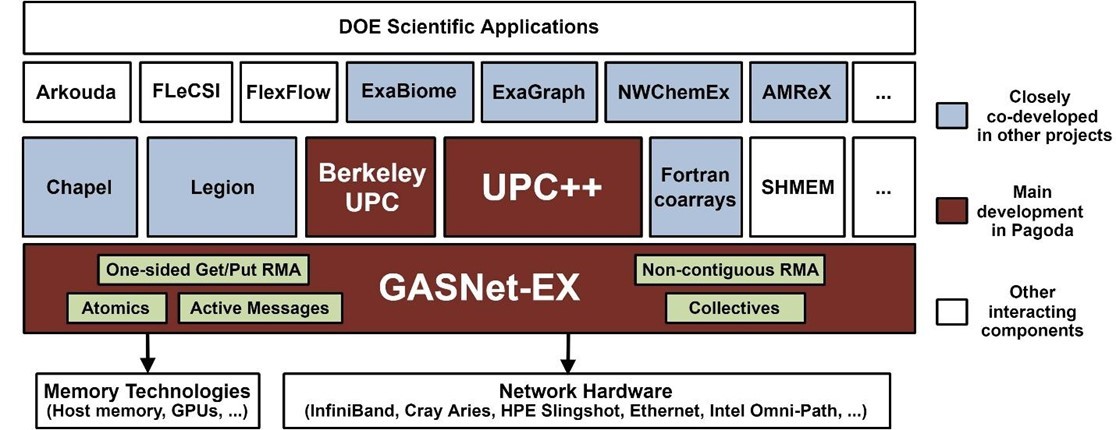
The Pagoda project software stack from application to networking and memory hardware. See https://go.lbl.gov/pagoda for more information. (Source)
GASNet-EX
GASNet-EX is an update to the 20-year-old GASNet-1 PGAS codebase and communication system. As part of this update, the GASNet interfaces have been redesigned to accommodate the emerging needs of exascale supercomputing and support communication services for a variety of PGAS programming models on current and future HPC architectures. This effort includes an implementation overhaul along with this major redesign of the software interfaces.
The motivating goals of this redesign, along with backwards compatibility for GASNet-1 clients include:
- Support more client asynchrony
- Enable more client adaptation
- Improve memory footprint
- Improve threading support
- Support offload to network hardware
- Support multi-client applications
- Support for device memory
Compatibility is important because GASNet has enjoyed adoption by a number of projects (Figure 2). The tight semantic match of GASNet-EX APIs to the client requirements and hardware capabilities often yields better performance than competing libraries due to its direct implementation over the native APIs for networks of interest in HPC.

GASNet adoption and portability.
Paul Hargrove, computer systems engineer 5 in the Applied Mathematics and Computational Research (AMCR) division at Lawrence Berkeley National Laboratory (Berkeley Lab) and PI of the ECP Pagoda project observed that the changes denoted by ‘-EX’ address how applications and HPC have changed over the last 20 years. For example, memory kinds are important to express how Remote Memory Accesses (RMA) now include gets and puts to/from GPU memory.
Organization and Programmer View
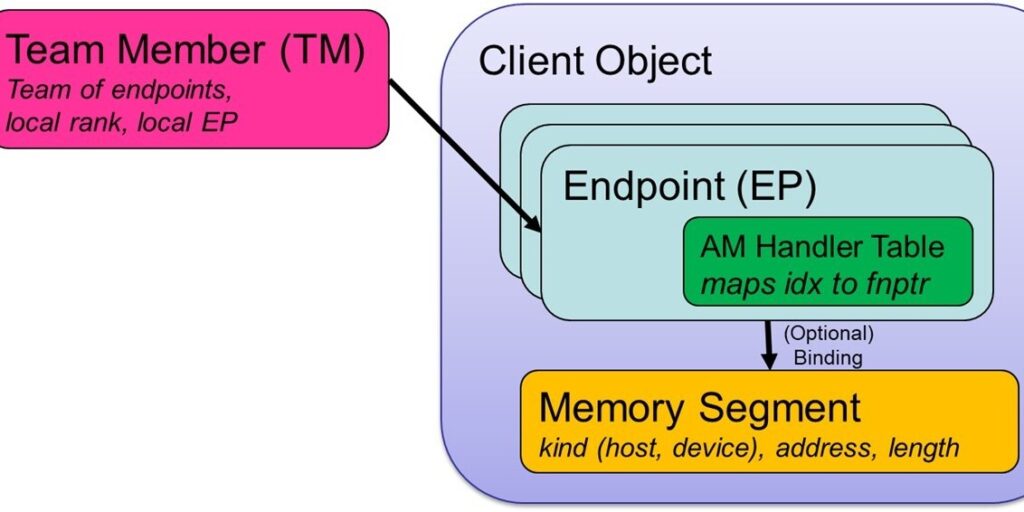
The Client Object resides at the top level of the GASNet-EX object hierarchy (Figure 3). It encompasses all of the interactions between a given programming model’s runtime system and GASNet. The term Client refers to the software layer sitting above GASNet and making GASNet calls, which creates this object at startup.
Each Client Object contains one or more Endpoints, which can be thought of as a lightweight communication context.
Each communication operation is generally associated with two or more endpoints. Each endpoint includes an active message (AM) handler table, which maps an active message index to the corresponding local function pointer. The endpoint also has an optional binding to a Segment object. A Segment object represents a contiguous region of local memory, which could be in the host DRAM or memory residing on a device like a GPU.
Endpoints can be grouped together into an ordered set of local and remote endpoints, which is a lightweight analogue of an MPI communicator. This group of endpoints are referred to as a Team Member (TM) and functions both as a mechanism for global naming of individual endpoints and for identifying the participants in a collective operation.
Most communication initiation operations in GASNet-EX take a TM argument, which implicitly specifies the client, endpoint, handlers and segment used by that operation.
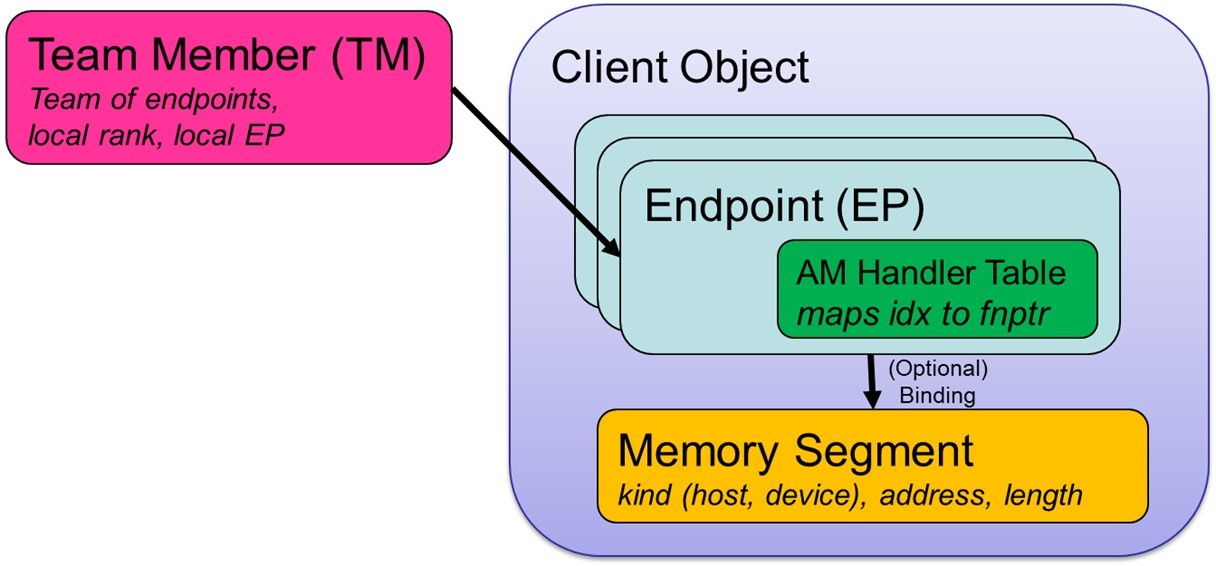
GASNet-EX high-level object model.
Hargrove continued, “GASNet-EX was created in acknowledgement that HPC systems and applications have gotten more complicated. Our users have found that computations that previously were very regular no longer exhibit such nice behavior. Communication of GPU-resident data is essential for performant heterogenous supercomputing. Giving users the ability to tie memory in a PGAS model to devices is important for efficiency and to provide computation/communication overlap and reduce time-to-solution. Asynchronous GPU put/get capabilities can help speed communications in a more natural way compared to MPI. Further, the team is working to increase scaling, eliminate locking, make threads first-class entities, and make use of endpoints explicit.
GASNet-EX was created in acknowledgement that HPC systems and applications have gotten more complicated. Our users have found that computations that previously were very regular no longer exhibit such nice behavior. Communication of GPU-resident data is essential for performant heterogenous supercomputing. Giving users the ability to tie memory in a PGAS model to devices is important for efficiency and to provide computation/communication overlap and reduce time-to-solution. Asynchronous GPU put/get capabilities can help speed communications in a more natural way compared to MPI. Further, the team is working to increase scaling, eliminate locking, make threads first-class entities, and make use of endpoints explicit. — Paul Hargrove, Berkeley Lab
UPC++

Paul Hargrove, AMCR, Berkeley Lab
UPC++ is the other half of the Pagoda story which is layered on top of GASNet-EX. “In particular”, Hargrove explained, “UPC++ generalizes the GASNet-EX communication operations for moving data around and to/from device memories. UPC++ is a higher-level abstraction supporting efficient one-sided communication for bulk data and rich C++ data types. UPC++ also provides the ability to easily move computation to data, which is often more natural for the programmer and/or more computationally efficient. UPC++ is an open-source template library that only requires a C++ compiler to use.
C++ Templated High-Level Abstractions
UPC++ provides high-level productivity abstractions appropriate for Partitioned Global Address Space (PGAS) programming such as: remote memory access (RMA), remote procedure call (RPC), support for accelerators (e.g., GPUs), and mechanisms for aggressive asynchrony to hide communication costs. UPC++ implements communication using GASNet-EX. It is designed to deliver high performance and portability from laptops to exascale supercomputers. HPC application software using UPC++ includes: MetaHipMer2 metagenome assembler, SIMCoV viral propagation simulation, NWChemEx TAMM, and graph computation kernels from ExaGraph.
UPC++ design characteristics are focused on:
- Wherever possible, UPC++ communication operations are asynchronous by default, to allow the overlap of computation and communication, and to encourage programmers to avoid global synchronization.
- All data motion is syntactically explicit, to encourage programmers to consider the cost of communication.
- UPC++ encourages the use of scalable data-structures and avoids non-scalable library features.
By default, all UPC++ communications are split-phased and begin with an initiate operation. The programmer is then free to perform whatever other work can be performed while the data is in transit, after which they perform a wait to ensure the data transfer operation has completed before proceeding (Figure 5).
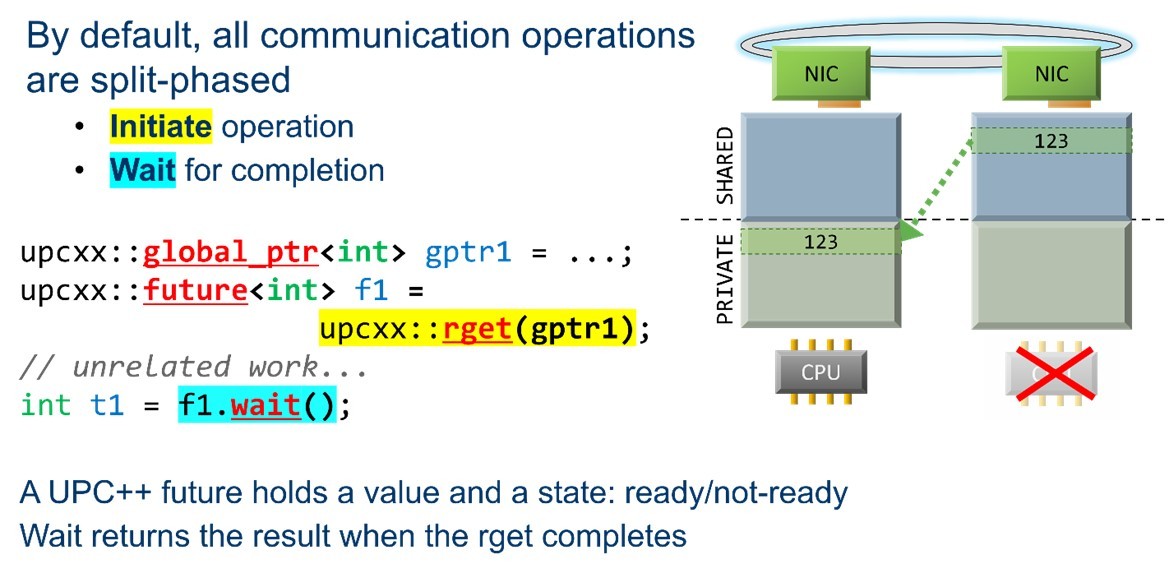
Example asynchronous RMA communication.
UPC++ exploits many features of modern C++ to help the programmer create understandable and maintainable code. Hargrove highlighted that, “UPC++ can be used in combination with C++ lambda expressions which has many programming advantages. With UPC++ and lambda expressions, the programmer can organize their source code so it specifies, at the exact location where UPC++ is required, the operations needed to move data to the computation or to move the computation to where the data resides.” This locality makes it much easier for people to understand the software and to maintain it.

Simple distributed hash table insertion using UPC++ Remote Procedure Call.
UPC++ enables the programmer to compose the powerful productivity features of the C++ standard template library seamlessly with remote procedure calls. For example Figure 6 demonstrates how one could easily implement a distributed hash table insertion operation by composing std::unordered_map with UPC++ RPC. In this example, each process holds its partition of the distributed hash table in the local_map variable (the key and value types are std::string for illustration purposes). The insertion operation uses a hash of the key to identify the partition corresponding to that key, and a UPC++ remote procedure call is sent to the owning process to perform the insertion on the appropriate copy of local_map. This entire operation is naturally asynchronous and returns a UPC++ future type, which the caller can later use to synchronize completion.
In addition to being highly productive, the UPC++ library can also deliver robust performance scalability on modern supercomputers. Figure 7 demonstrates the near-linear weak scaling on the NERSC Cori supercomputer of a slightly enhanced version of this distributed hash table. (For further details, see https://doi.org/10.25344/S4V88H).
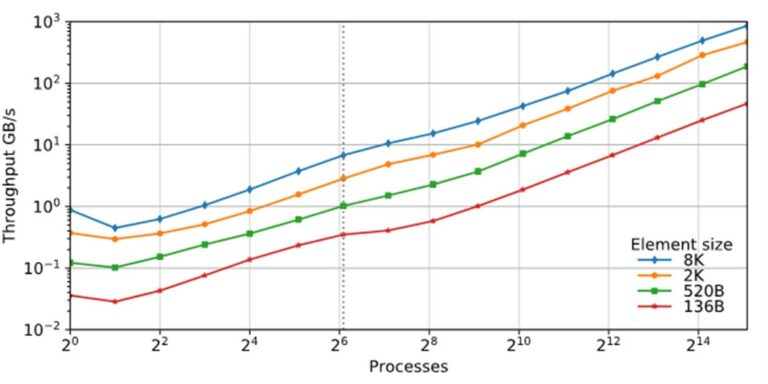
Performance of UPC++ distributed hash table insertion (weak scaling) for a variety of element sizes, using up to 34816 cores of NERSC Cori (Source).
For More Information
GASNet-EX (https://gasnet.lbl.gov/)
- Dan Bonachea, Paul H. Hargrove, “GASNet-EX: A High-Performance, Portable Communication Library for Exascale”,Languages and Compilers for Parallel Computing (LCPC’18), Salt Lake City, Utah, USA, October 11, 2018, LBNL-2001174, doi: 10.25344/S4QP4W
- 20th Anniversary Retrospective News Article: Linda Vu, “Berkeley Lab’s Networking Middleware GASNet Turns 20: Now, GASNet-EX is Gearing Up for the Exascale Era”,HPCWire (Lawrence Berkeley National Laboratory CS Area Communications), December 7, 2022, doi: 10.25344/S4BP4G
UPC++ (https://upcxx.lbl.gov/)
- John Bachan, Scott B. Baden, Steven Hofmeyr, Mathias Jacquelin, Amir Kamil, Dan Bonachea, Paul H. Hargrove, Hadia Ahmed. “UPC++: A High-Performance Communication Framework for Asynchronous Computation”, 33rd IEEE International Parallel & Distributed Processing Symposium (IPDPS’19), Rio de Janeiro, Brazil, IEEE, May 2019, doi: 10.25344/S4V88H
- SIMCoV, a simulation of lungs infected with Covid-19, written in UPC++ (journal paper)
- UPC++ SC21 tutorial: https://upcxx.lbl.gov/sc21
2023 Performance Results
GASNet-EX performance results are reported on the Berkeley Lab GASNet performance page for a wide range of machines.
Recent Frontier results measuring the performance of Get and Put RMA operations compared to the HPE Cray MPI library demonstrate that GASNet-EX can deliver comparable bandwidth performance using the world’s fastest supercomputer as of June 2023 (Figure 8).
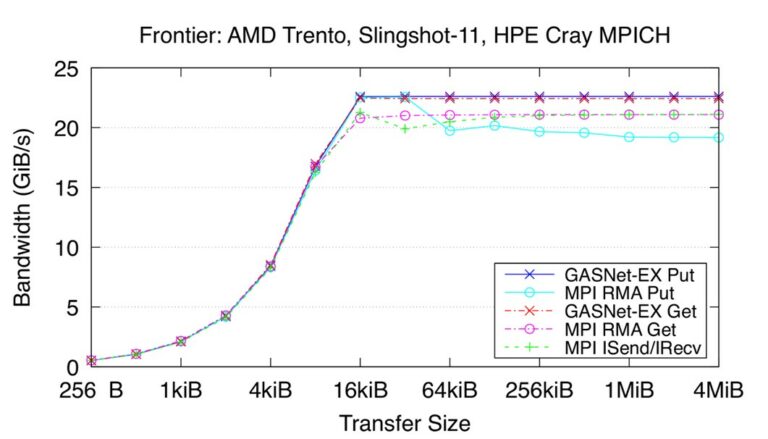
Recent bandwidth results for the Frontier HPE Cray EX Slingshot-11 Interconnect. Node config: 64-core 2 GHz AMD EPYC “Trento” 7453 CPUs, PE 8.3.3, GNU C 12.2.0, Cray MPICH 8.1.23, libfabric 1.15.2.0. (Source)
These “flood bandwidth” benchmark results measure the achievable interconnect bandwidth for a given transfer size by initiating a large number of non-blocking transfers (e.g., the “flood”) and waiting for them all to fully complete. The reported metric is the total volume of data transferred, divided by the total elapsed time. The benchmark reports unidirectional (one initiator to one target) flood bandwidths, where the passive target waits in an appropriate synchronization operation.[2]
Bandwidth tells only part of the story as many HPC applications can be limited by the latency of the communications operation. Figure 9 reports the mean time to fully complete a single round-trip RMA Put or Get operation on the Frontier Slingshot-11 interconnect, computed by timing a long sequence of blocking operations (lower is better). [3]
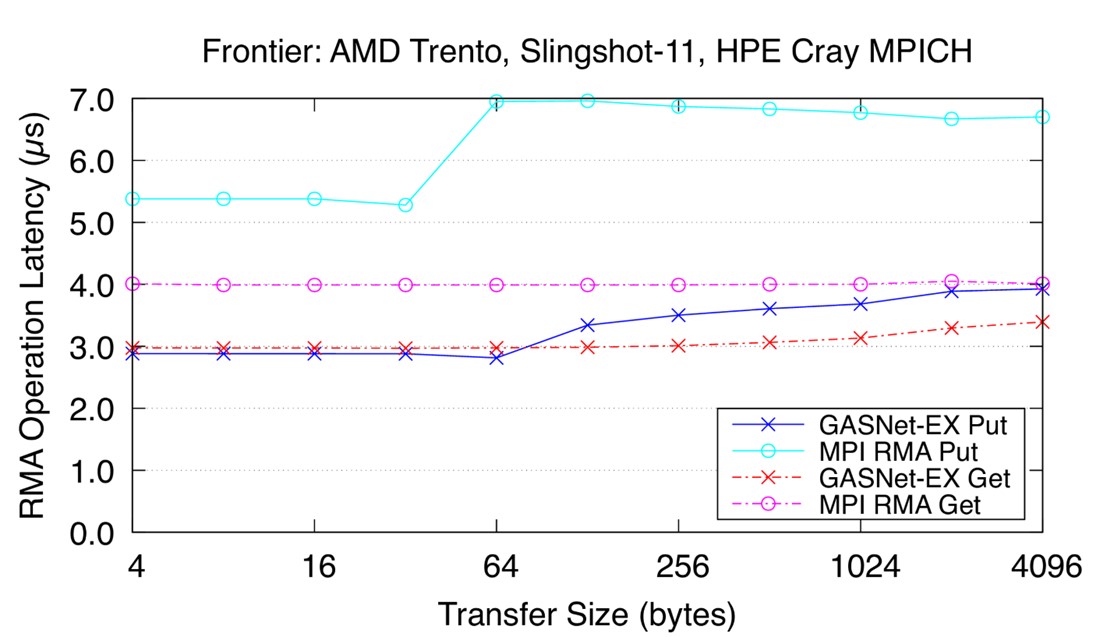
Recent latency results for the Frontier HPE Cray EX Slingshot-11 Interconnect. Node config: 64-core 2 GHz AMD EPYC “Trento” 7453 CPUs, PE 8.3.3, GNU C 12.2.0, Cray MPICH 8.1.23, libfabric 1.15.2.0. (Source)
Current GPU bandwidth and latency results on the Crusher testbed for the Frontier supercomputer are preliminary and a work in progress. [4]
Co-developed ECP projects
Hargrove noted that the Gordon Bell award finalist Exabiome Project is a closely co-developed project with Pagoda. Additional projects include ExaGraph, NWChemEx, and AMReX (Figure 1).
Kathy Yelick, senior advisor on computing and faculty affiliate at Berkeley Lab, PI of the Exascale Computing Project’s (ECP’s) Exabiome effort, and Vice Chancellor for Research at UC Berkeley, highlighted her team’s vision about the Exabiome project, “The Exabiome team is building a toolset to work with terabytes of data (containing trillions of base pairs) where we don’t understand all the proteins coded for by these base pairs at this time. Understanding the similarities between proteins and their interrelationships is important to understanding how microbiomes work. To do so requires very powerful computational tools that can infer the functions of related proteins in these massive datasets.” Microbiomes are cooperative communities of microbes. These organisms and the organisms in associated microbiomes are central players in climate change, environmental remediation, food production, and human health.[5]
To demonstrate the success of their approach, the Exabiome team utilized over 20,000 GPUs on Oak Ridge National Laboratory’s (ORNL’s) Summit supercomputer to perform a many-against-many protein similarity search on the largest available protein dataset. The performance observed was transformative because the search completed in hours rather than in weeks.
Achieving the GPU acceleration of HipMer and MetaHipMer2 to perform extreme-scale de novo genome assembly required the use of UPC++. MetaHipMer is a production metagenome assembler that was rewritten in UPC++ using ECP funding. It is tailored to metagenomics and optimized for GPUs.
The extraordinary (Gordon Bell Award submission worthy) performance benefit was highlighted in the 2023 ECP Annual Meeting presentation “ Exabiome: Then and Now” by Lenny Oliker (executive director of the Exabiome project, senior scientist, and group lead of the Berkeley Lab AMCR’s performance and algorithms group) and Kathy Yelick (senior advisor on computing and faculty affiliate at Berkeley Lab, PI of the ECP Exabiome effort, and Vice Chancellor for Research at UC Berkeley) notes that “the speedup from 2016 to 2021 is over 250× due to algorithmic improvements, use of GPUs and UPC++”.[6] The presentation also discussed the importance of the flood bandwidth performance (Figure 8). For more details see the “GASNet-EX: A High-Performance, Portable Communication Library for Exascale” and “GASNet-EX RMA Communication Performance on Recent Supercomputing Systems”.
The Exabiome website succinctly notes, “MetaHipMer’s high performance is based on several novel algorithmic advancements attained by leveraging the efficiency and programmability of the one-sided communication capabilities and RPC calls from UPC++, including optimized high-frequency k-mer analysis, communication-avoiding de Bruijn graph traversal, advanced I/O optimization, and extensive parallelization across the numerous and complex application phases.” [7]
A detailed discussion is provided in the article, “Accelerating large scale de novo metagenome assembly using GPUs” and associated presentation video.
Summary
The Pagoda Project community model addresses the software development needs of developers who use the PGAS globally shared address space to improve productivity and implement high-performance, scalable models. GPU acceleration increased the value of the PGAS model by making it easy to access data in a distributed heterogenous computing environment while embracing awareness of non-uniform communication costs, thus allowing optimization — even when data access behavior is too complex and irregular to be predictable.
This research was supported by the Exascale Computing Project (17-SC-20-SC), a joint project of the U.S. Department of Energy’s Office of Science and National Nuclear Security Administration, responsible for delivering a capable exascale ecosystem, including software, applications, and hardware technology, to support the nation’s exascale computing imperative.
Rob Farber is a global technology consultant and author with an extensive background in HPC and in developing machine learning technology that he applies at national laboratories and commercial organizations.
[2] https://doi.org/10.25344/S40C7D
[3] https://doi.org/10.25344/S40C7D
[4] See slide 22 for OLCF Crusher results at https://gasnet.lbl.gov/pubs/GASNet_PAW-ATM-22-Slides.pdf.
[5] https://www.exascaleproject.org/highlight/ Exabiome-gordon-bell-finalist-research-infers-the-functions-of-related-proteins-with-exascale-capable-homology-search/
[6] Slide