Using supercomputers, scientists are just starting to design proteins that self-assemble to combine and resemble life-giving molecules like hemoglobin.
Researchers are using HPC to design potentially life-saving proteins. In this TACC podcast, host Jorge Salazar discusses this groundbreaking work with Jens Glaser and Vyas Ramasubramani of the University of Michigan; and Anna Simon of UT Austin.
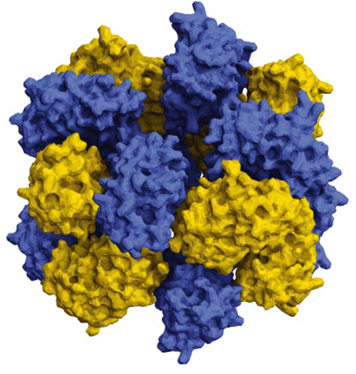
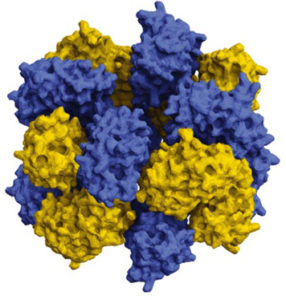
Using supercomputers, scientists are just starting to design proteins that self-assemble to combine and resemble life-giving molecules like hemoglobin. Hemoglobin molecules in red blood cells transport oxygen by changing their shape. Four copies of the same protein in hemoglobin open and close like flower petals, structurally coupled to respond to each other. The scientists say their methods could be applied to useful technologies such as pharmaceutical targeting, artificial energy harvesting, ‘smart’ sensing and building materials, and more.
The science team did this work by supercharging proteins, which means that they changed the subunits of proteins, the amino acids, to give the proteins an artificially high positive or negative charge. Using proteins derived from jellyfish, the scientists were able to assemble a complex sixteen protein structure composed of two stacked octamers by supercharging alone, findings that were reported in January of 2019 in the journal Nature Chemistry.
The team then used supercomputer simulations to validate and inform these experimental results. Supercomputer allocations on Stampede2 at the Texas Advanced Computing Center (TACC) and Comet at the San Diego Supercomputer Center (SDSC) were awarded to the researchers through XSEDE, the Extreme Science and Engineering Discovery Environment funded by the National Science Foundation (NSF).
We found that by taking proteins that don’t normally interact with each other, we can make copies that are either highly positively or highly negatively charged,” said study co-author Anna Simon, a postdoctoral researcher in the Ellington Lab of UT Austin. “Combining the highly positively and negatively charged copies, we can make the proteins assemble into very specific structured assemblies,” Simon said.
The scientists call their strategy ‘supercharged protein assembly,’ where they drive defined protein interactions by combining engineered supercharged variants.
We exploited a very well-known and basic principle from nature, that opposite charges attract,” added study co-author Jens Glaser. Glaser is an assistant research scientist in the Glotzer Group, Department of Chemical Engineering at the University of Michigan. “Anna Simon’s group found that when they mix these charged variants of green fluorescent protein, they get highly ordered structures. That was a real surprise,” Glaser said.