Challenges & Requirements
[SPONSORED GUEST ARTICLE] International Supercomputing Conference (ISC) is an annual conference that showcases the key technologies in the supercomputing field. It brings together experts from multiple specialist fields, including research into life sciences and genetics to shed light on the origins and mysteries in our lives. HPC, or high-performance computing, is making breakthroughs in supercomputing technologies and is catalyzing genome sequencing research.
The rapid development of supercomputing technologies causes a rise in the number of discovered genome sequences, presenting new challenges. Heavy data processing and parallel computing is necessary for homology search, alignment, and mutation detection processes, posing strict requirements on the data infrastructure of genomics data analysis platform.
Genome sequencing is the process of analyzing and determining the complete sequence of genes from blood or saliva. Its main phases cover extraction, analysis, and interpretation. Specifically, the analysis phase involves file format conversion, decompression, gene splicing, alignment, sequencing, deduplication, mutation detection, and joint genotyping. Its reliance on the performance of the bioinformatic analysis system makes it a main focus for High-Performance Computing (HPC) solutions for genome sequencing.
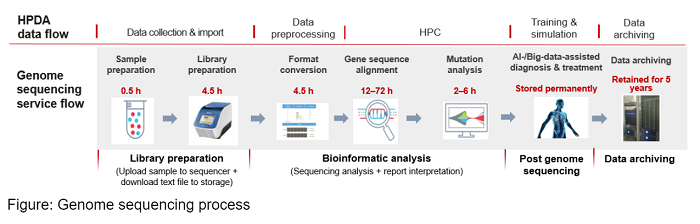
The second-gen whole genome sequencing (WGS) technologies are commonly used in coordination with a Genome Analysis Toolkit, or GATK, for bioinformatic analysis. Adjustments based on various services are required, such as adding quality control and filtering processes for mutation detection. The Burrows-Wheeler Alignment (BWA) tool is utilized to build indexes and conduct sequence alignment, Samtools is used for alignments sorting, and GATK is used to remove duplicate sequences, recalibrate base quality scores, and detect mutations.
Joint Innovation to Solve Industry Problems
As genome sequencing poses high requirements on data infrastructure, HPC solutions are required to provide high computing power and efficient storage to handle mass data. To this end, West China Hospital (WCH) called on the competitive advantages of Sailegene and Huawei to address these problems.
WCH is a leading body in multi-omics data analysis and genome application. It uses high-performance software algorithms to analyze running data, identify performance bottlenecks, design a top-level architecture, and build an acceleration analysis platform for multi-omics data. Sailegene leverages its years of expertise in GPU-accelerated bioinformation data analysis and the GPU/CPU heterogeneous parallel computing to accelerate genetic data analysis.
Huawei OceanStor Pacific scale-out storage supports advanced genetic data management systems, laying a high-performance storage foundation. Its key features and technologies are as follows:
- Fast performance: The single-thread bandwidth of private clients is 6 GB/s. BWA implements fast read and linear increase of aggregated bandwidth.
- Hybrid workload: Various HPC I/O models are supported.
- Flexible expansion: The system can be expanded to meet the demands of EB-scale genome research platform.
Thanks to the ultra-high single-thread write bandwidth, Huawei OceanStor Pacific can load mass data to the memory and process it during sequence alignment. Compared with the hospital’s legacy storage, OceanStor Pacific offers twice the single-thread read bandwidth and four times the single-thread write bandwidth. With just four nodes, it delivers an aggregate bandwidth of 30 GB/s read and 25 GB/s write, significantly enhancing the performance of the multi-omics joint innovation platform. Huawei OceanStor Pacific further analyzes service flows and I/O streams in different omics based on test data to optimize service processing.
From 24 Hours to 7 Minutes — A New Benchmark for Genome Sequencing
The three parties innovated to produce an acceleration analysis platform for multi-omics data that features new architectures, computing, and storage. It shortens the analysis time of a 30X human WGS germline mutation to just 7 minutes (down from 24 hours), meeting a wide range of precision medicine and big data needs in healthcare, and representing a significant breakthrough in medical research. Huawei OceanStor Pacific is in a prime position to drive innovation across the healthcare industry and help organizations in their journeys to an intelligent future.





