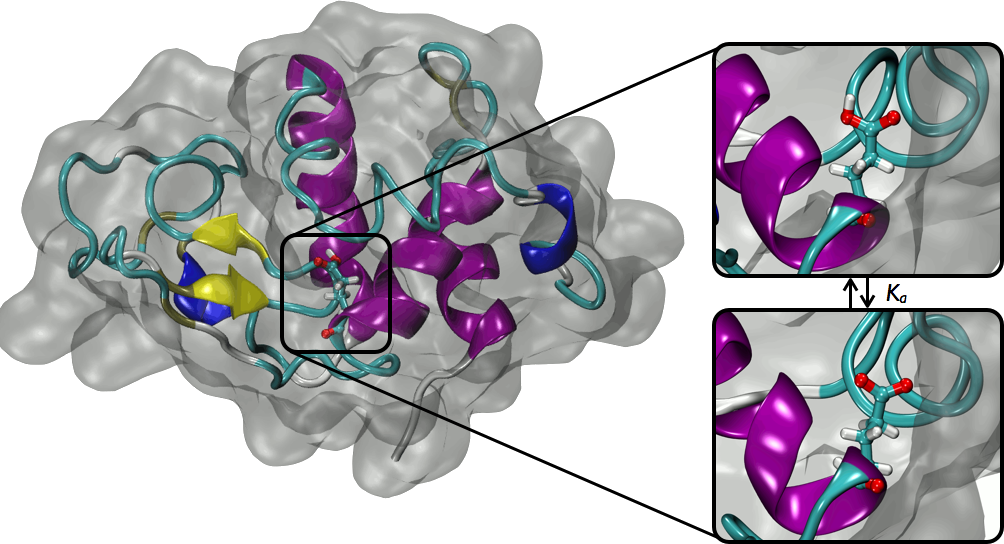
 The Amber Molecular Dynamics software is a well known and understood application for the structural dynamics of large biological molecules. With modern computer systems, a speedup in the computations can lead to studying events that occur on longer timescales, as well as statistical convergences. By incorporating the Intel Xeon Phi coprocessor into the workflow on a compute server, additional speedups can be obtained.
The Amber Molecular Dynamics software is a well known and understood application for the structural dynamics of large biological molecules. With modern computer systems, a speedup in the computations can lead to studying events that occur on longer timescales, as well as statistical convergences. By incorporating the Intel Xeon Phi coprocessor into the workflow on a compute server, additional speedups can be obtained.
A standard test setup was created with nodes comprising of the Intel Xeon E5-2697 v2 processor and the Intel Xeon Phi 7120P coprocessor. The theoretical peak performance of almost 3 Tflop/sec. Molecular dynamics happen in the range of femtoseconds, as such short time steps are needed to simulate the fastest vibrational motions. However, it is desirable to simulate up to microseconds, so billions of time steps are needed.
Amber basically solves a potential energy equation. Various optimization techniques can be used, including but not limited to reducing the transfer of data, removing locks, using MPI for load balancing, and paying attention to compiler optimization flags. The results of the work to port the code to use the Intel Xeon Phi coprocessor were exciting, in that a speedup over the original code, running on one node with 24 MPI tasks, compared to a 4 node system with 96 MPI tasks, was almost 2.9 X speedup.
While the performance in terms of peak teraflops was not achieved, further modifications to the code were identified for future research. This includes more vectorization to certain identified code segments.
Source: Intel, USA, San Diego Supercomputer Center, USA, University of California San Diego, USA
Turn Big Research into Big Results
Accelerate data analysis with Intel® Parallel Studio XE.
Try it >